| Full name: isocitrate dehydrogenase (NADP(+)) 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 15q26.1 | ||
| Entrez ID: 3418 | HGNC ID: HGNC:5383 | Ensembl Gene: ENSG00000182054 | OMIM ID: 147650 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of IDH2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IDH2 | 3418 | 210046_s_at | -0.0481 | 0.9144 | |
| GSE20347 | IDH2 | 3418 | 210046_s_at | -0.3937 | 0.1383 | |
| GSE23400 | IDH2 | 3418 | 210046_s_at | -0.3571 | 0.0001 | |
| GSE26886 | IDH2 | 3418 | 210046_s_at | -0.9080 | 0.0017 | |
| GSE29001 | IDH2 | 3418 | 210046_s_at | -0.4793 | 0.2665 | |
| GSE38129 | IDH2 | 3418 | 210046_s_at | -0.2915 | 0.2137 | |
| GSE45670 | IDH2 | 3418 | 210046_s_at | 0.0713 | 0.8512 | |
| GSE53622 | IDH2 | 3418 | 137610 | -0.6203 | 0.0000 | |
| GSE53624 | IDH2 | 3418 | 137610 | -0.5527 | 0.0000 | |
| GSE63941 | IDH2 | 3418 | 210046_s_at | 1.0815 | 0.2779 | |
| GSE77861 | IDH2 | 3418 | 210046_s_at | -0.5305 | 0.1032 | |
| GSE97050 | IDH2 | 3418 | A_23_P129209 | -0.5768 | 0.1659 | |
| SRP007169 | IDH2 | 3418 | RNAseq | -1.8369 | 0.0001 | |
| SRP008496 | IDH2 | 3418 | RNAseq | -1.4377 | 0.0000 | |
| SRP064894 | IDH2 | 3418 | RNAseq | -0.3832 | 0.0563 | |
| SRP133303 | IDH2 | 3418 | RNAseq | -0.5448 | 0.0003 | |
| SRP159526 | IDH2 | 3418 | RNAseq | -0.6439 | 0.0084 | |
| SRP193095 | IDH2 | 3418 | RNAseq | -0.7760 | 0.0000 | |
| SRP219564 | IDH2 | 3418 | RNAseq | -0.5400 | 0.2865 | |
| TCGA | IDH2 | 3418 | RNAseq | -0.3717 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
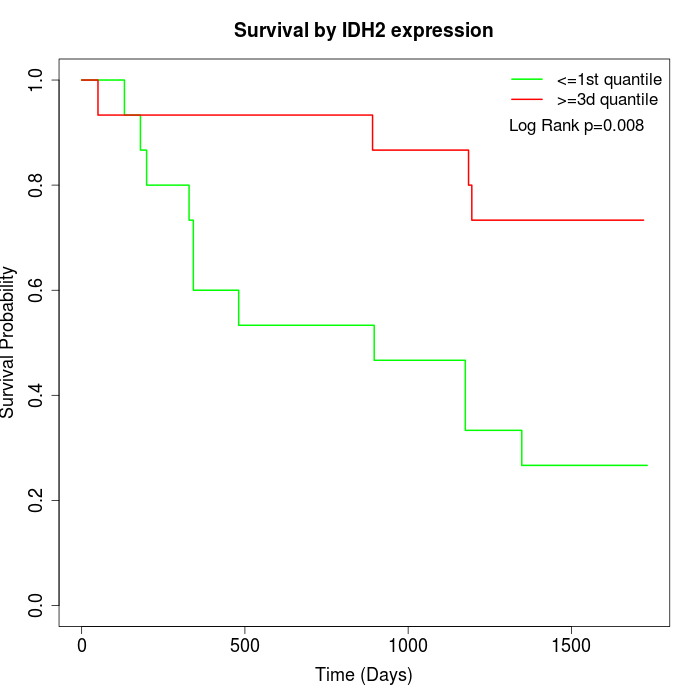
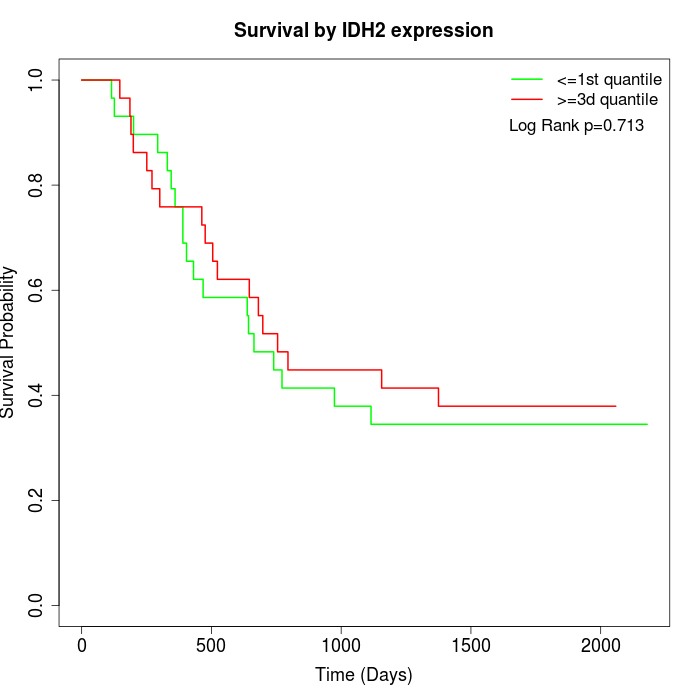
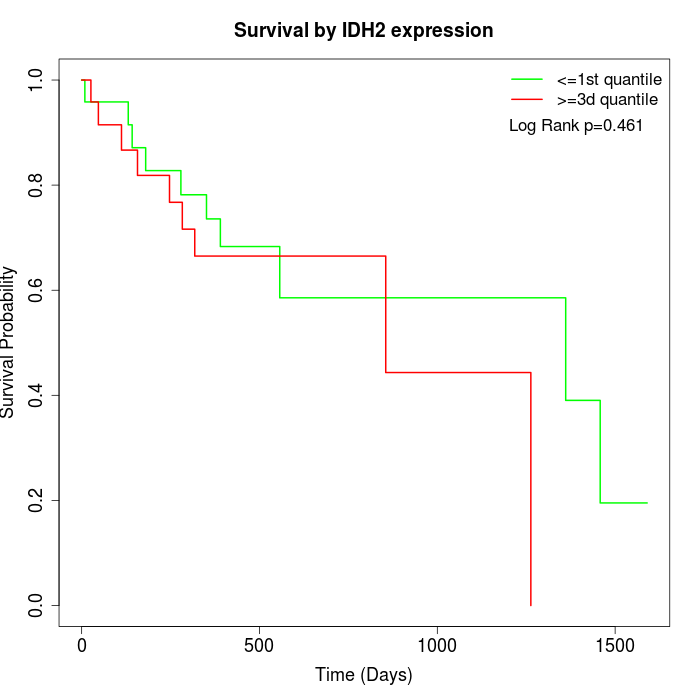
Survival by IDH2 expression:
Note: Click image to view full size file.
Copy number change of IDH2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IDH2 | 3418 | 8 | 3 | 19 | |
| GSE20123 | IDH2 | 3418 | 8 | 3 | 19 | |
| GSE43470 | IDH2 | 3418 | 5 | 4 | 34 | |
| GSE46452 | IDH2 | 3418 | 3 | 7 | 49 | |
| GSE47630 | IDH2 | 3418 | 8 | 11 | 21 | |
| GSE54993 | IDH2 | 3418 | 4 | 6 | 60 | |
| GSE54994 | IDH2 | 3418 | 7 | 6 | 40 | |
| GSE60625 | IDH2 | 3418 | 4 | 0 | 7 | |
| GSE74703 | IDH2 | 3418 | 4 | 3 | 29 | |
| GSE74704 | IDH2 | 3418 | 4 | 2 | 14 | |
| TCGA | IDH2 | 3418 | 18 | 12 | 66 |
Total number of gains: 73; Total number of losses: 57; Total Number of normals: 358.
Somatic mutations of IDH2:
Generating mutation plots.
Highly correlated genes for IDH2:
Showing top 20/314 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IDH2 | HDAC3 | 0.704691 | 3 | 0 | 3 |
| IDH2 | RASSF2 | 0.686925 | 3 | 0 | 3 |
| IDH2 | CD63 | 0.664254 | 3 | 0 | 3 |
| IDH2 | LYST | 0.658811 | 3 | 0 | 3 |
| IDH2 | DAPL1 | 0.65629 | 4 | 0 | 3 |
| IDH2 | MAP1LC3B | 0.648686 | 3 | 0 | 3 |
| IDH2 | CD99 | 0.647929 | 3 | 0 | 3 |
| IDH2 | EIF4H | 0.647638 | 4 | 0 | 4 |
| IDH2 | OAZ2 | 0.641214 | 4 | 0 | 3 |
| IDH2 | HINT3 | 0.637458 | 3 | 0 | 3 |
| IDH2 | GAB1 | 0.636979 | 4 | 0 | 3 |
| IDH2 | TPD52L2 | 0.636205 | 3 | 0 | 3 |
| IDH2 | COPA | 0.634914 | 3 | 0 | 3 |
| IDH2 | GRHPR | 0.624137 | 3 | 0 | 3 |
| IDH2 | ANKRD13A | 0.623143 | 5 | 0 | 4 |
| IDH2 | CCNG1 | 0.622919 | 4 | 0 | 3 |
| IDH2 | UQCRC2 | 0.616697 | 6 | 0 | 5 |
| IDH2 | COX7C | 0.614818 | 4 | 0 | 3 |
| IDH2 | SCUBE2 | 0.614089 | 3 | 0 | 3 |
| IDH2 | HERPUD1 | 0.613319 | 3 | 0 | 3 |
For details and further investigation, click here