| Full name: microtubule associated protein 1 light chain 3 beta | Alias Symbol: ATG8F | ||
| Type: protein-coding gene | Cytoband: 16q24.2 | ||
| Entrez ID: 81631 | HGNC ID: HGNC:13352 | Ensembl Gene: ENSG00000140941 | OMIM ID: 609604 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MAP1LC3B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP1LC3B | 81631 | 208786_s_at | -0.5297 | 0.3467 | |
| GSE20347 | MAP1LC3B | 81631 | 208786_s_at | -0.1405 | 0.4673 | |
| GSE23400 | MAP1LC3B | 81631 | 208786_s_at | -0.1876 | 0.0167 | |
| GSE26886 | MAP1LC3B | 81631 | 208786_s_at | -0.7402 | 0.0043 | |
| GSE29001 | MAP1LC3B | 81631 | 208786_s_at | -0.5578 | 0.1688 | |
| GSE38129 | MAP1LC3B | 81631 | 208786_s_at | -0.1867 | 0.2641 | |
| GSE45670 | MAP1LC3B | 81631 | 208786_s_at | -0.3286 | 0.0134 | |
| GSE53622 | MAP1LC3B | 81631 | 75134 | -0.2364 | 0.0017 | |
| GSE53624 | MAP1LC3B | 81631 | 75134 | -0.0295 | 0.7833 | |
| GSE63941 | MAP1LC3B | 81631 | 208786_s_at | -0.6434 | 0.3471 | |
| GSE77861 | MAP1LC3B | 81631 | 208786_s_at | 0.2324 | 0.2815 | |
| GSE97050 | MAP1LC3B | 81631 | A_32_P220715 | -0.3653 | 0.1421 | |
| SRP007169 | MAP1LC3B | 81631 | RNAseq | -1.0254 | 0.0458 | |
| SRP008496 | MAP1LC3B | 81631 | RNAseq | -0.7102 | 0.0141 | |
| SRP064894 | MAP1LC3B | 81631 | RNAseq | -0.3309 | 0.0844 | |
| SRP133303 | MAP1LC3B | 81631 | RNAseq | 0.4760 | 0.0170 | |
| SRP159526 | MAP1LC3B | 81631 | RNAseq | -0.2206 | 0.4643 | |
| SRP193095 | MAP1LC3B | 81631 | RNAseq | -0.3927 | 0.0074 | |
| SRP219564 | MAP1LC3B | 81631 | RNAseq | -0.5151 | 0.1277 | |
| TCGA | MAP1LC3B | 81631 | RNAseq | -0.0368 | 0.4217 |
Upregulated datasets: 0; Downregulated datasets: 1.
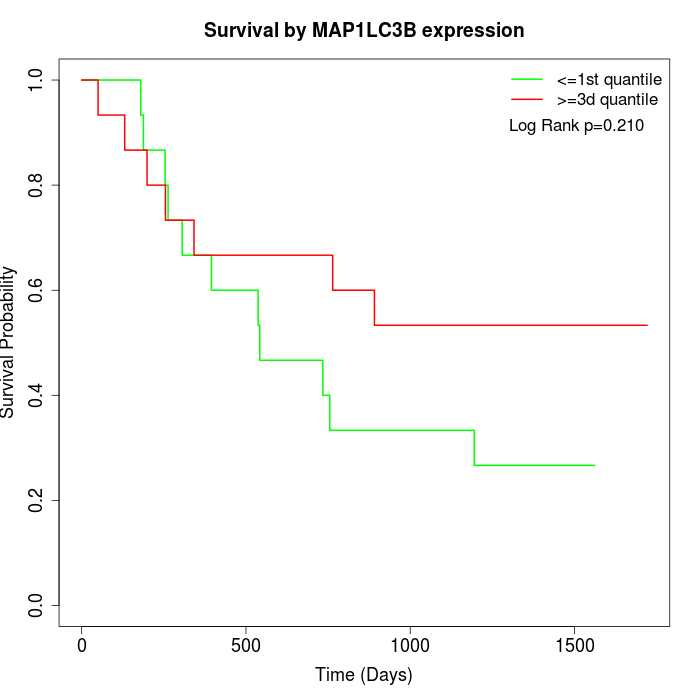
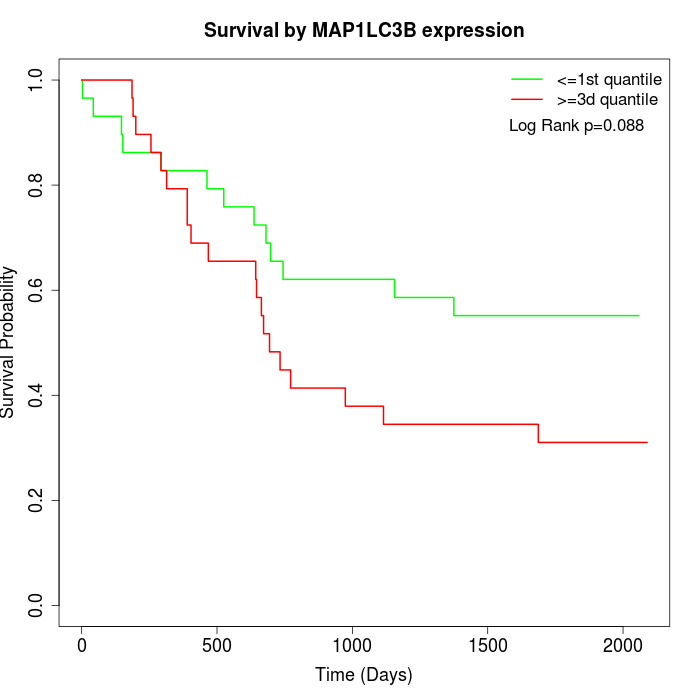
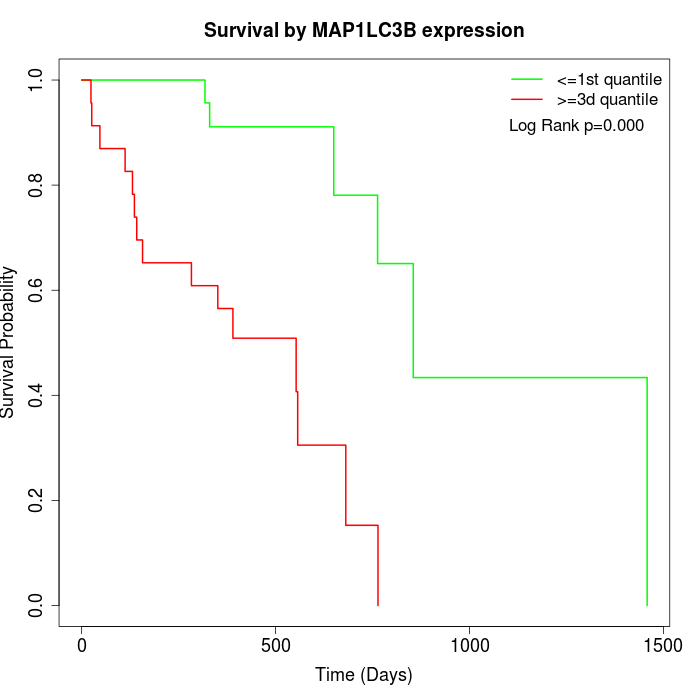
Survival by MAP1LC3B expression:
Note: Click image to view full size file.
Copy number change of MAP1LC3B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP1LC3B | 81631 | 4 | 2 | 24 | |
| GSE20123 | MAP1LC3B | 81631 | 4 | 2 | 24 | |
| GSE43470 | MAP1LC3B | 81631 | 1 | 13 | 29 | |
| GSE46452 | MAP1LC3B | 81631 | 38 | 1 | 20 | |
| GSE47630 | MAP1LC3B | 81631 | 11 | 9 | 20 | |
| GSE54993 | MAP1LC3B | 81631 | 3 | 4 | 63 | |
| GSE54994 | MAP1LC3B | 81631 | 9 | 11 | 33 | |
| GSE60625 | MAP1LC3B | 81631 | 4 | 0 | 7 | |
| GSE74703 | MAP1LC3B | 81631 | 1 | 9 | 26 | |
| GSE74704 | MAP1LC3B | 81631 | 3 | 2 | 15 | |
| TCGA | MAP1LC3B | 81631 | 27 | 15 | 54 |
Total number of gains: 105; Total number of losses: 68; Total Number of normals: 315.
Somatic mutations of MAP1LC3B:
Generating mutation plots.
Highly correlated genes for MAP1LC3B:
Showing top 20/437 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP1LC3B | MRPL10 | 0.787997 | 3 | 0 | 3 |
| MAP1LC3B | ARHGEF33 | 0.729747 | 3 | 0 | 3 |
| MAP1LC3B | ALDOC | 0.719627 | 3 | 0 | 3 |
| MAP1LC3B | SENP8 | 0.714643 | 3 | 0 | 3 |
| MAP1LC3B | ARL4C | 0.711579 | 3 | 0 | 3 |
| MAP1LC3B | RAB18 | 0.702403 | 3 | 0 | 3 |
| MAP1LC3B | PITPNM3 | 0.695371 | 3 | 0 | 3 |
| MAP1LC3B | ZNF398 | 0.688344 | 3 | 0 | 3 |
| MAP1LC3B | ZNF546 | 0.682438 | 4 | 0 | 3 |
| MAP1LC3B | VDAC2 | 0.680637 | 5 | 0 | 4 |
| MAP1LC3B | ECSCR | 0.678555 | 3 | 0 | 3 |
| MAP1LC3B | HDAC3 | 0.675091 | 3 | 0 | 3 |
| MAP1LC3B | PDK2 | 0.674778 | 3 | 0 | 3 |
| MAP1LC3B | PPP2CA | 0.672687 | 4 | 0 | 4 |
| MAP1LC3B | PTGR1 | 0.672582 | 3 | 0 | 3 |
| MAP1LC3B | UGP2 | 0.669529 | 8 | 0 | 7 |
| MAP1LC3B | VPS29 | 0.667628 | 5 | 0 | 4 |
| MAP1LC3B | PCMTD2 | 0.665851 | 3 | 0 | 3 |
| MAP1LC3B | SKAP2 | 0.664261 | 5 | 0 | 3 |
| MAP1LC3B | OGFRL1 | 0.663208 | 3 | 0 | 3 |
For details and further investigation, click here