| Full name: integrin subunit alpha 2 | Alias Symbol: CD49b | ||
| Type: protein-coding gene | Cytoband: 5q11.2 | ||
| Entrez ID: 3673 | HGNC ID: HGNC:6137 | Ensembl Gene: ENSG00000164171 | OMIM ID: 192974 |
| Drug and gene relationship at DGIdb | |||
ITGA2 involved pathways:
Expression of ITGA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGA2 | 3673 | 227314_at | -0.5657 | 0.3731 | |
| GSE20347 | ITGA2 | 3673 | 205032_at | -0.1103 | 0.5801 | |
| GSE23400 | ITGA2 | 3673 | 205032_at | -0.0590 | 0.3593 | |
| GSE26886 | ITGA2 | 3673 | 227314_at | 0.4260 | 0.2060 | |
| GSE29001 | ITGA2 | 3673 | 205032_at | -0.2570 | 0.2956 | |
| GSE38129 | ITGA2 | 3673 | 205032_at | 0.1562 | 0.5105 | |
| GSE45670 | ITGA2 | 3673 | 227314_at | 0.3349 | 0.2827 | |
| GSE53622 | ITGA2 | 3673 | 47368 | 0.4684 | 0.0021 | |
| GSE53624 | ITGA2 | 3673 | 36450 | -0.0598 | 0.6083 | |
| GSE63941 | ITGA2 | 3673 | 227314_at | -0.1320 | 0.8758 | |
| GSE77861 | ITGA2 | 3673 | 227314_at | -0.0443 | 0.9394 | |
| GSE97050 | ITGA2 | 3673 | A_32_P178800 | 0.8214 | 0.1316 | |
| SRP007169 | ITGA2 | 3673 | RNAseq | 0.8070 | 0.0579 | |
| SRP008496 | ITGA2 | 3673 | RNAseq | 0.1657 | 0.6707 | |
| SRP064894 | ITGA2 | 3673 | RNAseq | -0.0911 | 0.7922 | |
| SRP133303 | ITGA2 | 3673 | RNAseq | 0.5315 | 0.1062 | |
| SRP159526 | ITGA2 | 3673 | RNAseq | -0.2634 | 0.4706 | |
| SRP193095 | ITGA2 | 3673 | RNAseq | -0.0017 | 0.9934 | |
| SRP219564 | ITGA2 | 3673 | RNAseq | -0.3052 | 0.6672 | |
| TCGA | ITGA2 | 3673 | RNAseq | 0.5313 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
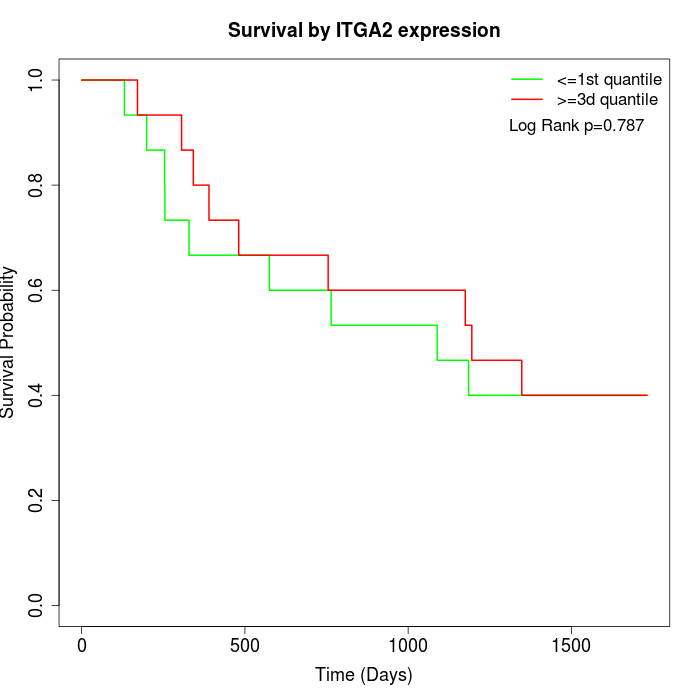
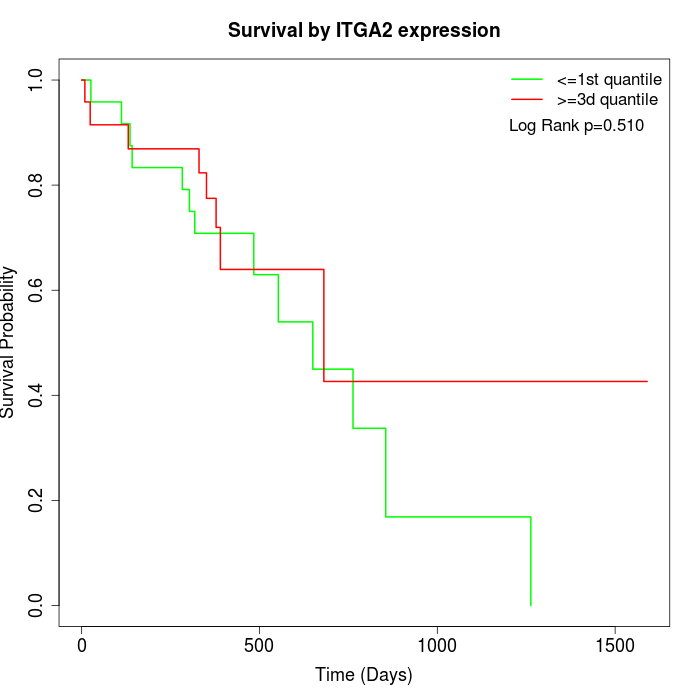
Survival by ITGA2 expression:
Note: Click image to view full size file.
Copy number change of ITGA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGA2 | 3673 | 1 | 12 | 17 | |
| GSE20123 | ITGA2 | 3673 | 1 | 12 | 17 | |
| GSE43470 | ITGA2 | 3673 | 1 | 12 | 30 | |
| GSE46452 | ITGA2 | 3673 | 1 | 27 | 31 | |
| GSE47630 | ITGA2 | 3673 | 2 | 16 | 22 | |
| GSE54993 | ITGA2 | 3673 | 8 | 1 | 61 | |
| GSE54994 | ITGA2 | 3673 | 5 | 17 | 31 | |
| GSE60625 | ITGA2 | 3673 | 0 | 0 | 11 | |
| GSE74703 | ITGA2 | 3673 | 1 | 9 | 26 | |
| GSE74704 | ITGA2 | 3673 | 1 | 5 | 14 | |
| TCGA | ITGA2 | 3673 | 10 | 45 | 41 |
Total number of gains: 31; Total number of losses: 156; Total Number of normals: 301.
Somatic mutations of ITGA2:
Generating mutation plots.
Highly correlated genes for ITGA2:
Showing top 20/240 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGA2 | RNGTT | 0.774455 | 3 | 0 | 3 |
| ITGA2 | JMJD1C | 0.770815 | 3 | 0 | 3 |
| ITGA2 | AP3B1 | 0.738495 | 3 | 0 | 3 |
| ITGA2 | GLCE | 0.728089 | 3 | 0 | 3 |
| ITGA2 | GAPVD1 | 0.722745 | 3 | 0 | 3 |
| ITGA2 | FBXO3 | 0.713515 | 3 | 0 | 3 |
| ITGA2 | DNASE1L1 | 0.712541 | 4 | 0 | 3 |
| ITGA2 | CPVL | 0.708358 | 3 | 0 | 3 |
| ITGA2 | ASXL2 | 0.704573 | 4 | 0 | 4 |
| ITGA2 | BLOC1S2 | 0.700239 | 3 | 0 | 3 |
| ITGA2 | LMAN2 | 0.698722 | 3 | 0 | 3 |
| ITGA2 | GSTZ1 | 0.696131 | 3 | 0 | 3 |
| ITGA2 | LY75 | 0.693848 | 4 | 0 | 4 |
| ITGA2 | WDR41 | 0.69361 | 4 | 0 | 4 |
| ITGA2 | DNAJB14 | 0.691377 | 3 | 0 | 3 |
| ITGA2 | METTL9 | 0.687604 | 3 | 0 | 3 |
| ITGA2 | CLPX | 0.687205 | 3 | 0 | 3 |
| ITGA2 | TBCK | 0.686721 | 3 | 0 | 3 |
| ITGA2 | MAP4K5 | 0.686708 | 3 | 0 | 3 |
| ITGA2 | MFAP3 | 0.686042 | 3 | 0 | 3 |
For details and further investigation, click here