| Full name: lectin, mannose binding 2 | Alias Symbol: GP36B|VIP36 | ||
| Type: protein-coding gene | Cytoband: 5q35.3 | ||
| Entrez ID: 10960 | HGNC ID: HGNC:16986 | Ensembl Gene: ENSG00000169223 | OMIM ID: 609551 |
| Drug and gene relationship at DGIdb | |||
Expression of LMAN2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMAN2 | 10960 | 200805_at | -0.3826 | 0.1802 | |
| GSE20347 | LMAN2 | 10960 | 200805_at | -0.8955 | 0.0000 | |
| GSE23400 | LMAN2 | 10960 | 200805_at | -0.5441 | 0.0000 | |
| GSE26886 | LMAN2 | 10960 | 200805_at | -1.0919 | 0.0000 | |
| GSE29001 | LMAN2 | 10960 | 200805_at | -0.6447 | 0.0111 | |
| GSE38129 | LMAN2 | 10960 | 200805_at | -0.4451 | 0.0047 | |
| GSE45670 | LMAN2 | 10960 | 200805_at | -0.3300 | 0.0447 | |
| GSE53622 | LMAN2 | 10960 | 94206 | -0.4644 | 0.0000 | |
| GSE53624 | LMAN2 | 10960 | 94206 | -0.6389 | 0.0000 | |
| GSE63941 | LMAN2 | 10960 | 200805_at | -0.2818 | 0.3832 | |
| GSE77861 | LMAN2 | 10960 | 200805_at | -0.7114 | 0.0097 | |
| GSE97050 | LMAN2 | 10960 | A_24_P9285 | 0.2025 | 0.6797 | |
| SRP007169 | LMAN2 | 10960 | RNAseq | -2.1094 | 0.0000 | |
| SRP008496 | LMAN2 | 10960 | RNAseq | -1.9761 | 0.0000 | |
| SRP064894 | LMAN2 | 10960 | RNAseq | 0.0171 | 0.9537 | |
| SRP133303 | LMAN2 | 10960 | RNAseq | -0.4125 | 0.0081 | |
| SRP159526 | LMAN2 | 10960 | RNAseq | -0.6627 | 0.0482 | |
| SRP193095 | LMAN2 | 10960 | RNAseq | -0.8368 | 0.0000 | |
| SRP219564 | LMAN2 | 10960 | RNAseq | 0.0736 | 0.8482 | |
| TCGA | LMAN2 | 10960 | RNAseq | -0.0013 | 0.9782 |
Upregulated datasets: 0; Downregulated datasets: 3.
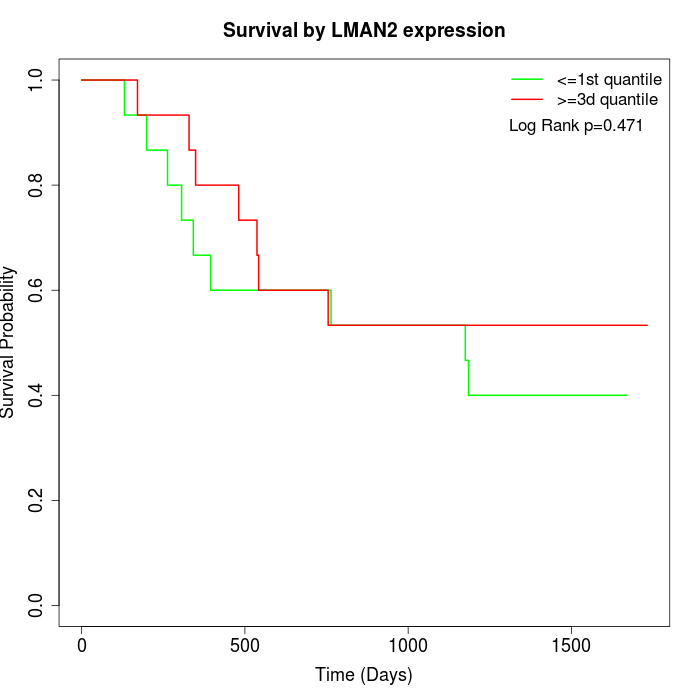
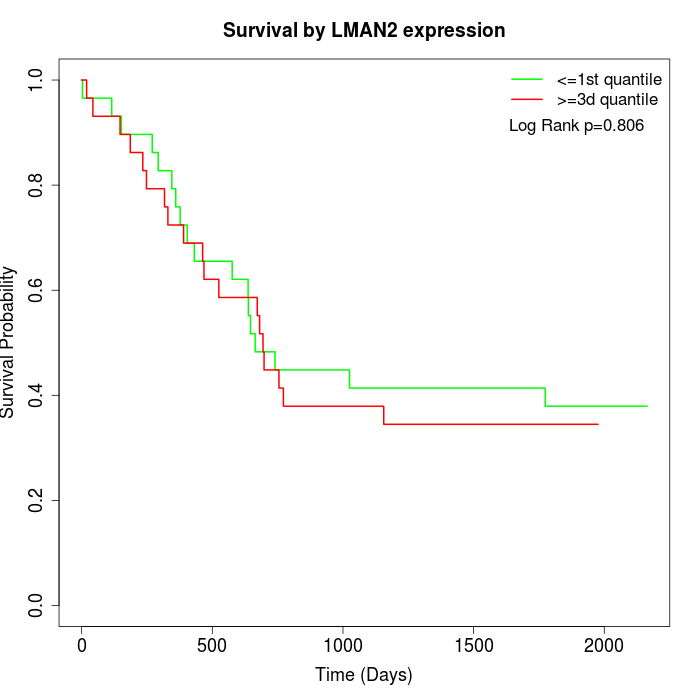
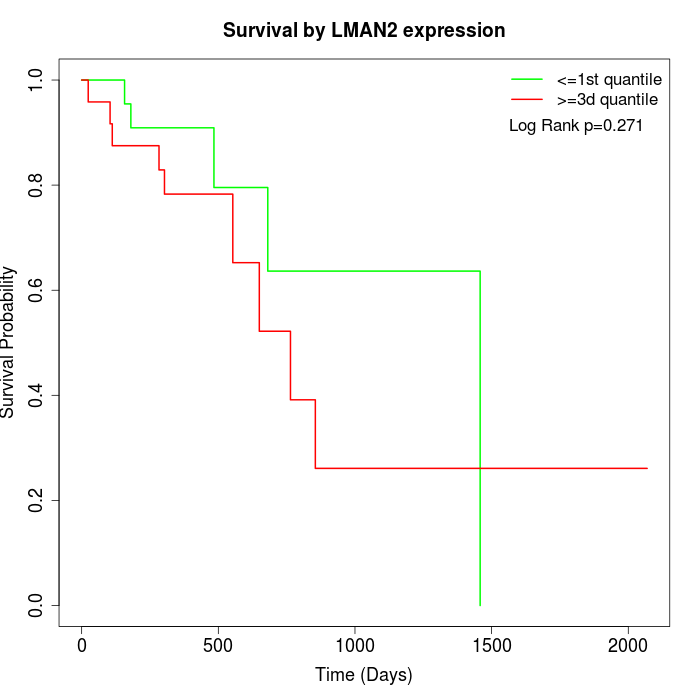
Survival by LMAN2 expression:
Note: Click image to view full size file.
Copy number change of LMAN2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMAN2 | 10960 | 2 | 13 | 15 | |
| GSE20123 | LMAN2 | 10960 | 2 | 13 | 15 | |
| GSE43470 | LMAN2 | 10960 | 1 | 11 | 31 | |
| GSE46452 | LMAN2 | 10960 | 0 | 27 | 32 | |
| GSE47630 | LMAN2 | 10960 | 1 | 20 | 19 | |
| GSE54993 | LMAN2 | 10960 | 10 | 2 | 58 | |
| GSE54994 | LMAN2 | 10960 | 2 | 17 | 34 | |
| GSE60625 | LMAN2 | 10960 | 1 | 0 | 10 | |
| GSE74703 | LMAN2 | 10960 | 1 | 7 | 28 | |
| GSE74704 | LMAN2 | 10960 | 1 | 6 | 13 | |
| TCGA | LMAN2 | 10960 | 8 | 34 | 54 |
Total number of gains: 29; Total number of losses: 150; Total Number of normals: 309.
Somatic mutations of LMAN2:
Generating mutation plots.
Highly correlated genes for LMAN2:
Showing top 20/1433 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMAN2 | GRN | 0.782185 | 9 | 0 | 9 |
| LMAN2 | SIAE | 0.758775 | 5 | 0 | 5 |
| LMAN2 | NAGK | 0.758607 | 12 | 0 | 11 |
| LMAN2 | AMER2 | 0.744326 | 3 | 0 | 3 |
| LMAN2 | HERPUD1 | 0.742407 | 3 | 0 | 3 |
| LMAN2 | TECR | 0.741518 | 10 | 0 | 9 |
| LMAN2 | RAB25 | 0.736799 | 11 | 0 | 11 |
| LMAN2 | BLNK | 0.735842 | 11 | 0 | 11 |
| LMAN2 | SLC25A39 | 0.732661 | 4 | 0 | 4 |
| LMAN2 | NAPA | 0.730424 | 10 | 0 | 10 |
| LMAN2 | ORMDL2 | 0.72899 | 9 | 0 | 9 |
| LMAN2 | CIAPIN1 | 0.726855 | 3 | 0 | 3 |
| LMAN2 | PPP1R7 | 0.726725 | 10 | 0 | 9 |
| LMAN2 | RER1 | 0.723355 | 10 | 0 | 10 |
| LMAN2 | CXCR2 | 0.721236 | 10 | 0 | 10 |
| LMAN2 | SMPD2 | 0.71901 | 11 | 0 | 11 |
| LMAN2 | RMND5B | 0.711657 | 11 | 0 | 10 |
| LMAN2 | GALE | 0.709666 | 11 | 0 | 11 |
| LMAN2 | IRX3 | 0.708309 | 4 | 0 | 4 |
| LMAN2 | MAML2 | 0.707703 | 3 | 0 | 3 |
For details and further investigation, click here