| Full name: inositol-trisphosphate 3-kinase C | Alias Symbol: IP3KC|IP3-3KC | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 80271 | HGNC ID: HGNC:14897 | Ensembl Gene: ENSG00000086544 | OMIM ID: 606476 |
| Drug and gene relationship at DGIdb | |||
ITPKC involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway |
Expression of ITPKC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITPKC | 80271 | 213076_at | -0.3018 | 0.6742 | |
| GSE20347 | ITPKC | 80271 | 213076_at | -0.9792 | 0.0001 | |
| GSE23400 | ITPKC | 80271 | 213076_at | -0.7804 | 0.0000 | |
| GSE26886 | ITPKC | 80271 | 213076_at | -1.4710 | 0.0000 | |
| GSE29001 | ITPKC | 80271 | 213076_at | -1.2430 | 0.0069 | |
| GSE38129 | ITPKC | 80271 | 213076_at | -0.7857 | 0.0009 | |
| GSE45670 | ITPKC | 80271 | 213076_at | -0.3463 | 0.2623 | |
| GSE53622 | ITPKC | 80271 | 129788 | -1.3608 | 0.0000 | |
| GSE53624 | ITPKC | 80271 | 129788 | -1.4382 | 0.0000 | |
| GSE63941 | ITPKC | 80271 | 213076_at | -0.0845 | 0.8485 | |
| GSE77861 | ITPKC | 80271 | 213076_at | -0.7053 | 0.0316 | |
| GSE97050 | ITPKC | 80271 | A_24_P202567 | -0.5499 | 0.2151 | |
| SRP007169 | ITPKC | 80271 | RNAseq | -2.6254 | 0.0000 | |
| SRP008496 | ITPKC | 80271 | RNAseq | -2.8436 | 0.0000 | |
| SRP064894 | ITPKC | 80271 | RNAseq | -1.9682 | 0.0000 | |
| SRP133303 | ITPKC | 80271 | RNAseq | -0.9103 | 0.0002 | |
| SRP159526 | ITPKC | 80271 | RNAseq | -1.7605 | 0.0000 | |
| SRP193095 | ITPKC | 80271 | RNAseq | -1.4584 | 0.0000 | |
| SRP219564 | ITPKC | 80271 | RNAseq | -1.1215 | 0.0061 | |
| TCGA | ITPKC | 80271 | RNAseq | 0.0317 | 0.6982 |
Upregulated datasets: 0; Downregulated datasets: 10.
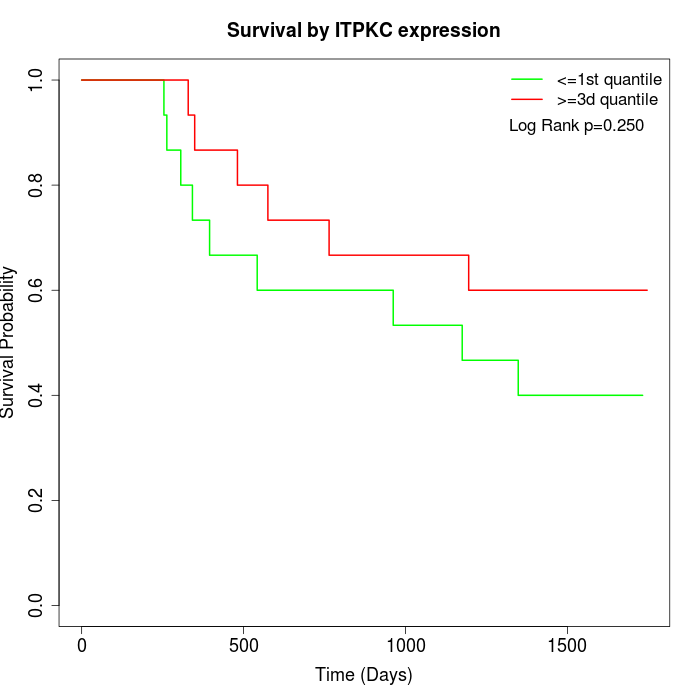
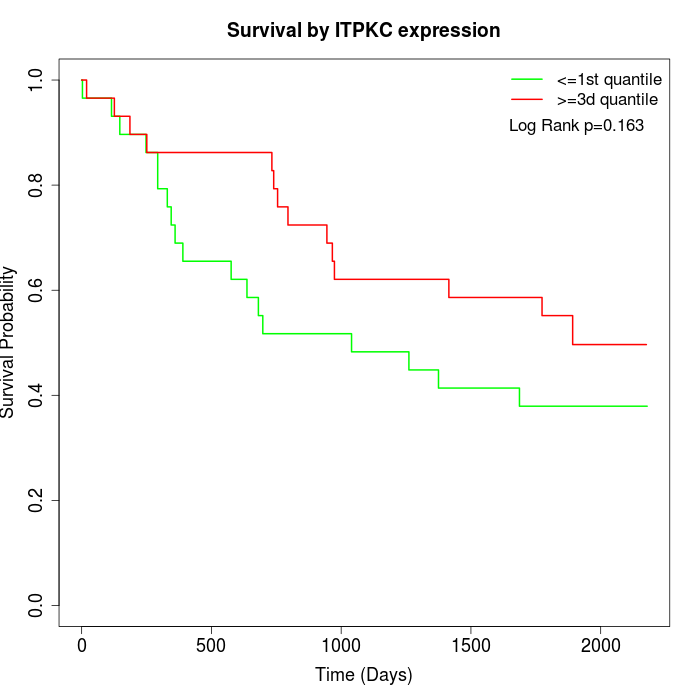
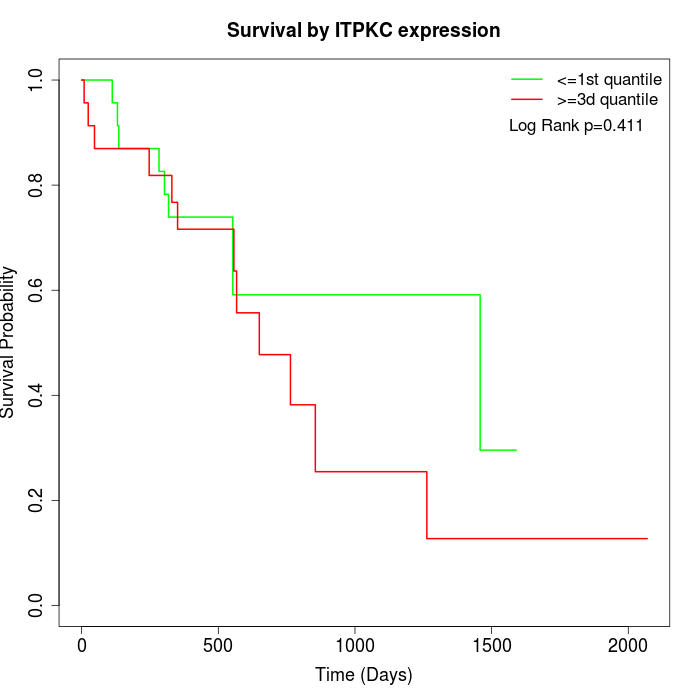
Survival by ITPKC expression:
Note: Click image to view full size file.
Copy number change of ITPKC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITPKC | 80271 | 6 | 5 | 19 | |
| GSE20123 | ITPKC | 80271 | 6 | 4 | 20 | |
| GSE43470 | ITPKC | 80271 | 3 | 9 | 31 | |
| GSE46452 | ITPKC | 80271 | 48 | 1 | 10 | |
| GSE47630 | ITPKC | 80271 | 9 | 5 | 26 | |
| GSE54993 | ITPKC | 80271 | 17 | 4 | 49 | |
| GSE54994 | ITPKC | 80271 | 8 | 9 | 36 | |
| GSE60625 | ITPKC | 80271 | 9 | 0 | 2 | |
| GSE74703 | ITPKC | 80271 | 3 | 6 | 27 | |
| GSE74704 | ITPKC | 80271 | 6 | 2 | 12 | |
| TCGA | ITPKC | 80271 | 14 | 16 | 66 |
Total number of gains: 129; Total number of losses: 61; Total Number of normals: 298.
Somatic mutations of ITPKC:
Generating mutation plots.
Highly correlated genes for ITPKC:
Showing top 20/1604 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITPKC | CTTNBP2NL | 0.809412 | 6 | 0 | 6 |
| ITPKC | CPEB4 | 0.790192 | 7 | 0 | 7 |
| ITPKC | PIM1 | 0.786881 | 10 | 0 | 10 |
| ITPKC | CA13 | 0.780899 | 3 | 0 | 3 |
| ITPKC | APOB | 0.778478 | 4 | 0 | 4 |
| ITPKC | MALL | 0.777278 | 9 | 0 | 9 |
| ITPKC | JOSD2 | 0.770687 | 3 | 0 | 3 |
| ITPKC | ACAA1 | 0.769569 | 9 | 0 | 9 |
| ITPKC | CCNYL1 | 0.768927 | 4 | 0 | 4 |
| ITPKC | TMEM79 | 0.764977 | 7 | 0 | 6 |
| ITPKC | MPZL3 | 0.763621 | 6 | 0 | 6 |
| ITPKC | GRHL3 | 0.76181 | 5 | 0 | 5 |
| ITPKC | FCHO2 | 0.759252 | 6 | 0 | 6 |
| ITPKC | LYPD3 | 0.758178 | 10 | 0 | 10 |
| ITPKC | FRMD4B | 0.754904 | 10 | 0 | 10 |
| ITPKC | NLRX1 | 0.748377 | 9 | 0 | 9 |
| ITPKC | LACTBL1 | 0.743013 | 3 | 0 | 3 |
| ITPKC | CSTB | 0.741624 | 9 | 0 | 9 |
| ITPKC | ANXA1 | 0.740747 | 9 | 0 | 9 |
| ITPKC | C5orf66-AS1 | 0.739976 | 5 | 0 | 5 |
For details and further investigation, click here