| Full name: glucagon | Alias Symbol: GLP1|GLP2|GRPP|GLP-1 | ||
| Type: protein-coding gene | Cytoband: 2q24.2 | ||
| Entrez ID: 2641 | HGNC ID: HGNC:4191 | Ensembl Gene: ENSG00000115263 | OMIM ID: 138030 |
| Drug and gene relationship at DGIdb | |||
GCG involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04911 | Insulin secretion | |
| hsa04922 | Glucagon signaling pathway |
Expression of GCG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GCG | 2641 | 206422_at | -0.0308 | 0.9297 | |
| GSE20347 | GCG | 2641 | 206422_at | -0.0624 | 0.2145 | |
| GSE23400 | GCG | 2641 | 206422_at | -0.1022 | 0.0001 | |
| GSE26886 | GCG | 2641 | 206422_at | 0.0591 | 0.4805 | |
| GSE29001 | GCG | 2641 | 206422_at | -0.0357 | 0.7821 | |
| GSE38129 | GCG | 2641 | 206422_at | -0.1539 | 0.0234 | |
| GSE45670 | GCG | 2641 | 206422_at | 0.0101 | 0.8919 | |
| GSE53622 | GCG | 2641 | 14063 | -0.0505 | 0.5146 | |
| GSE53624 | GCG | 2641 | 14063 | -0.0229 | 0.8205 | |
| GSE63941 | GCG | 2641 | 206422_at | 0.0376 | 0.7923 | |
| GSE77861 | GCG | 2641 | 206422_at | -0.0398 | 0.6798 | |
| TCGA | GCG | 2641 | RNAseq | -1.9549 | 0.3624 |
Upregulated datasets: 0; Downregulated datasets: 0.
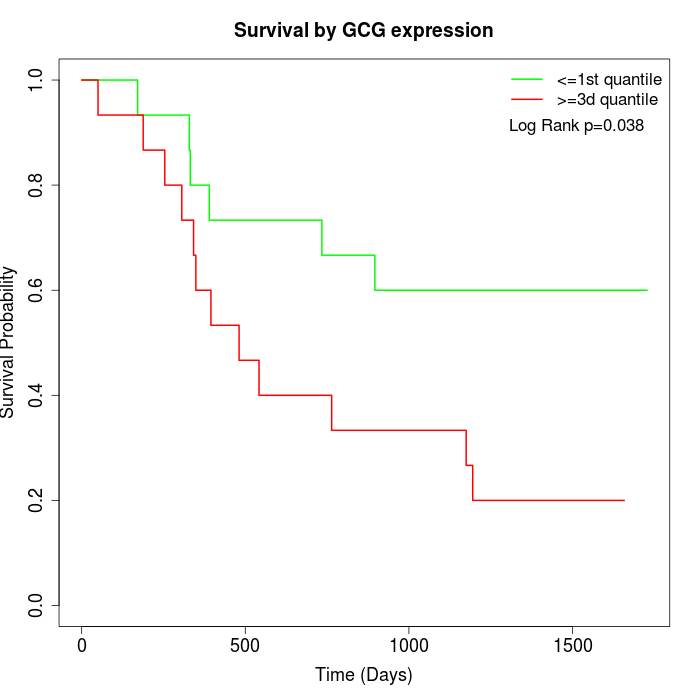
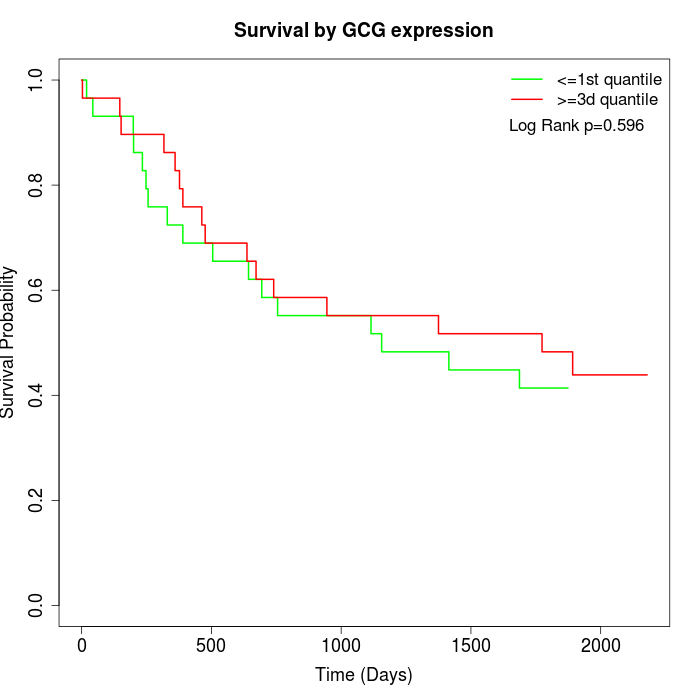
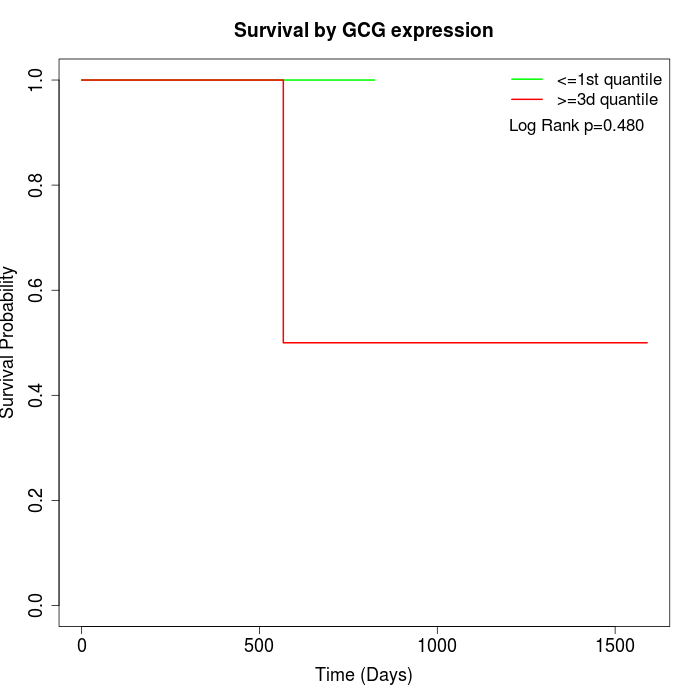
Survival by GCG expression:
Note: Click image to view full size file.
Copy number change of GCG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GCG | 2641 | 4 | 1 | 25 | |
| GSE20123 | GCG | 2641 | 4 | 1 | 25 | |
| GSE43470 | GCG | 2641 | 4 | 1 | 38 | |
| GSE46452 | GCG | 2641 | 1 | 4 | 54 | |
| GSE47630 | GCG | 2641 | 5 | 3 | 32 | |
| GSE54993 | GCG | 2641 | 0 | 5 | 65 | |
| GSE54994 | GCG | 2641 | 11 | 2 | 40 | |
| GSE60625 | GCG | 2641 | 0 | 3 | 8 | |
| GSE74703 | GCG | 2641 | 3 | 1 | 32 | |
| GSE74704 | GCG | 2641 | 3 | 0 | 17 | |
| TCGA | GCG | 2641 | 23 | 10 | 63 |
Total number of gains: 58; Total number of losses: 31; Total Number of normals: 399.
Somatic mutations of GCG:
Generating mutation plots.
Highly correlated genes for GCG:
Showing top 20/267 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GCG | KCNA4 | 0.725394 | 3 | 0 | 3 |
| GCG | HPCAL4 | 0.715766 | 4 | 0 | 4 |
| GCG | PSG6 | 0.702944 | 3 | 0 | 3 |
| GCG | BCAN | 0.70068 | 3 | 0 | 3 |
| GCG | KCNJ10 | 0.685749 | 3 | 0 | 3 |
| GCG | SERPINC1 | 0.68037 | 3 | 0 | 3 |
| GCG | MYOD1 | 0.674157 | 4 | 0 | 4 |
| GCG | IL17A | 0.673946 | 3 | 0 | 3 |
| GCG | TMPRSS15 | 0.668952 | 4 | 0 | 4 |
| GCG | IFNA8 | 0.667388 | 3 | 0 | 3 |
| GCG | PMFBP1 | 0.665538 | 3 | 0 | 3 |
| GCG | KCNAB3 | 0.658427 | 3 | 0 | 3 |
| GCG | B3GAT1 | 0.658176 | 3 | 0 | 3 |
| GCG | NTSR2 | 0.651678 | 3 | 0 | 3 |
| GCG | CFHR5 | 0.650983 | 3 | 0 | 3 |
| GCG | GC | 0.644432 | 3 | 0 | 3 |
| GCG | ULBP1 | 0.643492 | 4 | 0 | 3 |
| GCG | AQP8 | 0.642987 | 4 | 0 | 3 |
| GCG | PDZD3 | 0.639396 | 3 | 0 | 3 |
| GCG | FBP2 | 0.633112 | 4 | 0 | 3 |
For details and further investigation, click here