| Full name: potassium voltage-gated channel subfamily B member 2 | Alias Symbol: Kv2.2 | ||
| Type: protein-coding gene | Cytoband: 8q21.11 | ||
| Entrez ID: 9312 | HGNC ID: HGNC:6232 | Ensembl Gene: ENSG00000182674 | OMIM ID: 607738 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNB2 | 9312 | 208172_s_at | -0.0190 | 0.9458 | |
| GSE20347 | KCNB2 | 9312 | 208172_s_at | -0.0167 | 0.8951 | |
| GSE23400 | KCNB2 | 9312 | 208172_s_at | -0.1929 | 0.0000 | |
| GSE26886 | KCNB2 | 9312 | 208172_s_at | -0.3398 | 0.0118 | |
| GSE29001 | KCNB2 | 9312 | 208172_s_at | -0.1178 | 0.4606 | |
| GSE38129 | KCNB2 | 9312 | 208172_s_at | -0.1714 | 0.1952 | |
| GSE45670 | KCNB2 | 9312 | 208172_s_at | -0.0163 | 0.8962 | |
| GSE53622 | KCNB2 | 9312 | 115850 | -0.3474 | 0.0554 | |
| GSE53624 | KCNB2 | 9312 | 115850 | -0.2279 | 0.0653 | |
| GSE63941 | KCNB2 | 9312 | 208172_s_at | 0.3538 | 0.0947 | |
| GSE77861 | KCNB2 | 9312 | 208172_s_at | -0.0701 | 0.5647 | |
| GSE97050 | KCNB2 | 9312 | A_33_P3415350 | -0.8502 | 0.1934 | |
| TCGA | KCNB2 | 9312 | RNAseq | -0.6777 | 0.2785 |
Upregulated datasets: 0; Downregulated datasets: 0.
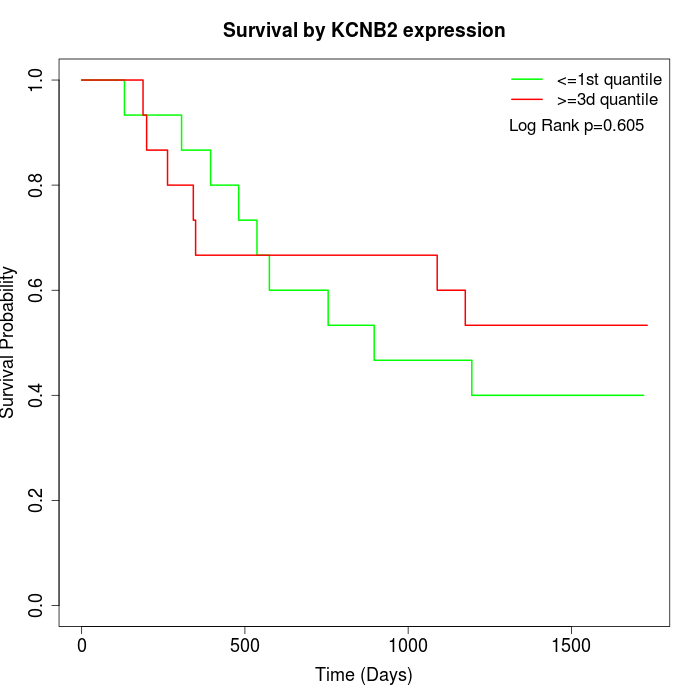
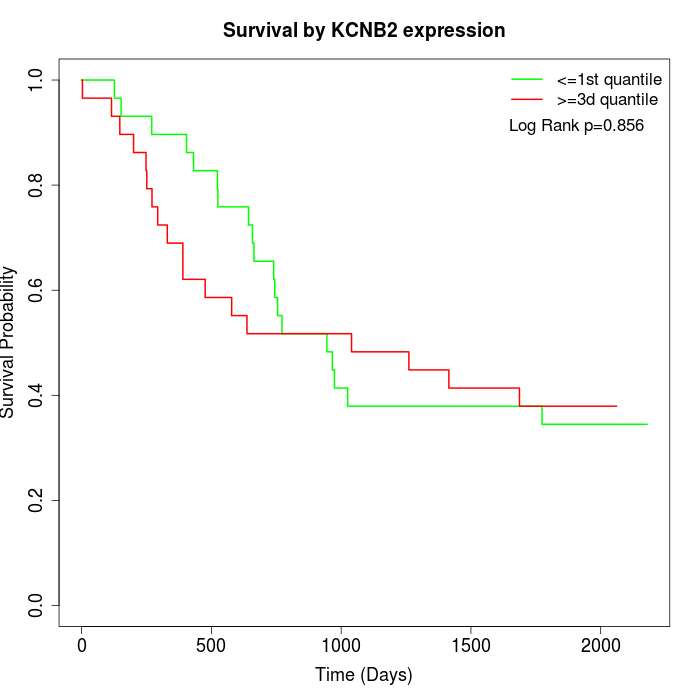
Survival by KCNB2 expression:
Note: Click image to view full size file.
Copy number change of KCNB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNB2 | 9312 | 15 | 1 | 14 | |
| GSE20123 | KCNB2 | 9312 | 16 | 1 | 13 | |
| GSE43470 | KCNB2 | 9312 | 18 | 1 | 24 | |
| GSE46452 | KCNB2 | 9312 | 21 | 1 | 37 | |
| GSE47630 | KCNB2 | 9312 | 23 | 0 | 17 | |
| GSE54993 | KCNB2 | 9312 | 0 | 19 | 51 | |
| GSE54994 | KCNB2 | 9312 | 30 | 0 | 23 | |
| GSE60625 | KCNB2 | 9312 | 0 | 4 | 7 | |
| GSE74703 | KCNB2 | 9312 | 15 | 1 | 20 | |
| GSE74704 | KCNB2 | 9312 | 11 | 0 | 9 | |
| TCGA | KCNB2 | 9312 | 51 | 2 | 43 |
Total number of gains: 200; Total number of losses: 30; Total Number of normals: 258.
Somatic mutations of KCNB2:
Generating mutation plots.
Highly correlated genes for KCNB2:
Showing top 20/761 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNB2 | LINC00563 | 0.690262 | 4 | 0 | 4 |
| KCNB2 | RET | 0.687748 | 5 | 0 | 5 |
| KCNB2 | UBQLN3 | 0.682403 | 6 | 0 | 5 |
| KCNB2 | TAS2R16 | 0.682268 | 4 | 0 | 4 |
| KCNB2 | SHISA9 | 0.67649 | 3 | 0 | 3 |
| KCNB2 | GAD2 | 0.671158 | 6 | 0 | 5 |
| KCNB2 | DNAH9 | 0.670514 | 6 | 0 | 6 |
| KCNB2 | GRM6 | 0.666175 | 6 | 0 | 6 |
| KCNB2 | IL1RAPL2 | 0.665482 | 5 | 0 | 4 |
| KCNB2 | STMN4 | 0.664352 | 4 | 0 | 4 |
| KCNB2 | IHH | 0.663263 | 5 | 0 | 4 |
| KCNB2 | FSHR | 0.660158 | 5 | 0 | 4 |
| KCNB2 | TCP10L | 0.659861 | 5 | 0 | 4 |
| KCNB2 | IFNA17 | 0.658562 | 6 | 0 | 5 |
| KCNB2 | DRD5 | 0.657992 | 4 | 0 | 3 |
| KCNB2 | CCL16 | 0.657498 | 6 | 0 | 5 |
| KCNB2 | USH2A | 0.656355 | 3 | 0 | 3 |
| KCNB2 | NCR1 | 0.656034 | 7 | 0 | 6 |
| KCNB2 | GYPB | 0.655725 | 5 | 0 | 5 |
| KCNB2 | CST8 | 0.654643 | 6 | 0 | 6 |
For details and further investigation, click here