| Full name: potassium voltage-gated channel modifier subfamily G member 2 | Alias Symbol: Kv6.2|KCNF2 | ||
| Type: protein-coding gene | Cytoband: 18q23 | ||
| Entrez ID: 26251 | HGNC ID: HGNC:6249 | Ensembl Gene: ENSG00000178342 | OMIM ID: 605696 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNG2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNG2 | 26251 | 208550_x_at | -0.7590 | 0.0540 | |
| GSE20347 | KCNG2 | 26251 | 208550_x_at | -0.5878 | 0.0000 | |
| GSE23400 | KCNG2 | 26251 | 208550_x_at | -0.2238 | 0.0000 | |
| GSE26886 | KCNG2 | 26251 | 208550_x_at | -0.1162 | 0.4213 | |
| GSE29001 | KCNG2 | 26251 | 208550_x_at | -0.4252 | 0.0005 | |
| GSE38129 | KCNG2 | 26251 | 208550_x_at | -0.6103 | 0.0002 | |
| GSE45670 | KCNG2 | 26251 | 208550_x_at | -0.4059 | 0.0382 | |
| GSE53622 | KCNG2 | 26251 | 38790 | -0.7386 | 0.0000 | |
| GSE53624 | KCNG2 | 26251 | 38790 | -1.1411 | 0.0000 | |
| GSE63941 | KCNG2 | 26251 | 208550_x_at | 0.0286 | 0.9075 | |
| GSE77861 | KCNG2 | 26251 | 208550_x_at | -0.3808 | 0.0449 | |
| GSE97050 | KCNG2 | 26251 | A_23_P61171 | -0.2034 | 0.4929 | |
| TCGA | KCNG2 | 26251 | RNAseq | -0.0354 | 0.9200 |
Upregulated datasets: 0; Downregulated datasets: 1.
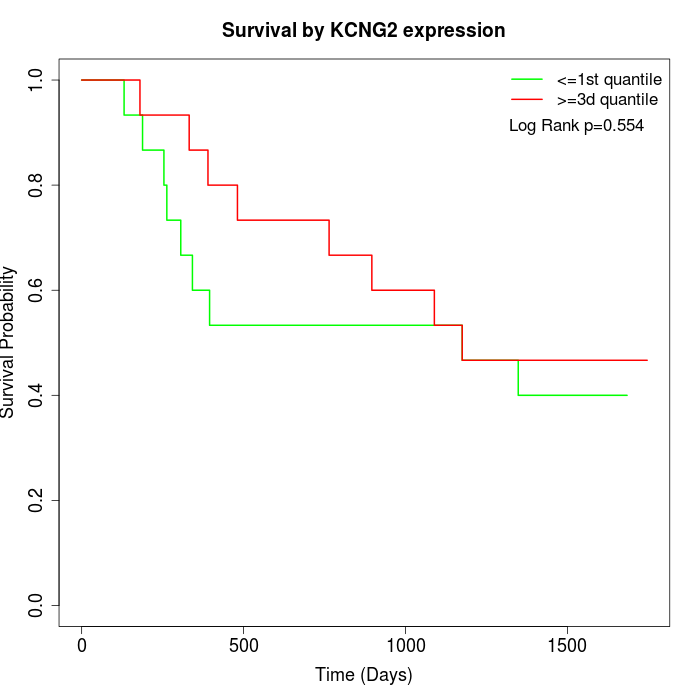
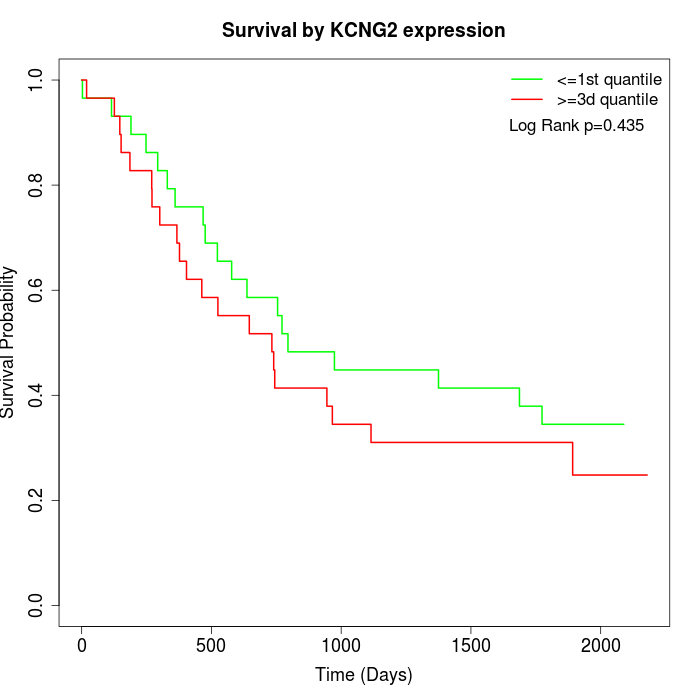
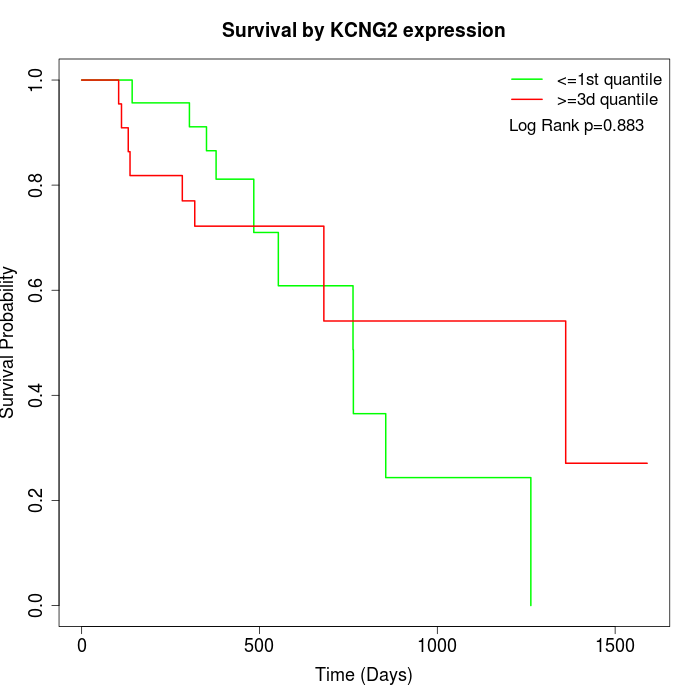
Survival by KCNG2 expression:
Note: Click image to view full size file.
Copy number change of KCNG2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNG2 | 26251 | 1 | 12 | 17 | |
| GSE20123 | KCNG2 | 26251 | 1 | 12 | 17 | |
| GSE43470 | KCNG2 | 26251 | 0 | 12 | 31 | |
| GSE46452 | KCNG2 | 26251 | 1 | 26 | 32 | |
| GSE47630 | KCNG2 | 26251 | 5 | 22 | 13 | |
| GSE54993 | KCNG2 | 26251 | 9 | 0 | 61 | |
| GSE54994 | KCNG2 | 26251 | 1 | 19 | 33 | |
| GSE60625 | KCNG2 | 26251 | 0 | 4 | 7 | |
| GSE74703 | KCNG2 | 26251 | 0 | 8 | 28 | |
| GSE74704 | KCNG2 | 26251 | 0 | 9 | 11 | |
| TCGA | KCNG2 | 26251 | 7 | 45 | 44 |
Total number of gains: 25; Total number of losses: 169; Total Number of normals: 294.
Somatic mutations of KCNG2:
Generating mutation plots.
Highly correlated genes for KCNG2:
Showing top 20/1561 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNG2 | APIP | 0.743199 | 3 | 0 | 3 |
| KCNG2 | AGGF1 | 0.739918 | 3 | 0 | 3 |
| KCNG2 | DOCK9 | 0.739349 | 9 | 0 | 9 |
| KCNG2 | RAB5A | 0.738669 | 9 | 0 | 9 |
| KCNG2 | CHIC2 | 0.734869 | 6 | 0 | 5 |
| KCNG2 | DCAKD | 0.734843 | 3 | 0 | 3 |
| KCNG2 | EPB41L4A | 0.733849 | 6 | 0 | 6 |
| KCNG2 | MTPN | 0.727245 | 5 | 0 | 5 |
| KCNG2 | TRIP11 | 0.725767 | 3 | 0 | 3 |
| KCNG2 | TMCO4 | 0.722263 | 3 | 0 | 3 |
| KCNG2 | CETN3 | 0.722085 | 4 | 0 | 4 |
| KCNG2 | TMEM40 | 0.721048 | 9 | 0 | 8 |
| KCNG2 | MIR210HG | 0.718948 | 5 | 0 | 5 |
| KCNG2 | TP53I3 | 0.718236 | 9 | 0 | 8 |
| KCNG2 | THRB | 0.716646 | 4 | 0 | 4 |
| KCNG2 | MAP2K3 | 0.714351 | 3 | 0 | 3 |
| KCNG2 | RNF146 | 0.712759 | 3 | 0 | 3 |
| KCNG2 | GBP6 | 0.712302 | 3 | 0 | 3 |
| KCNG2 | A2ML1 | 0.711558 | 5 | 0 | 5 |
| KCNG2 | MAPK3 | 0.710242 | 9 | 0 | 9 |
For details and further investigation, click here