| Full name: mitogen-activated protein kinase kinase 3 | Alias Symbol: MEK3|MKK3|MAPKK3 | ||
| Type: protein-coding gene | Cytoband: 17p11.2 | ||
| Entrez ID: 5606 | HGNC ID: HGNC:6843 | Ensembl Gene: ENSG00000034152 | OMIM ID: 602315 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAP2K3 involved pathways:
Expression of MAP2K3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | MAP2K3 | 5606 | 52733 | -0.4861 | 0.0000 | |
| GSE53624 | MAP2K3 | 5606 | 52733 | -0.5079 | 0.0000 | |
| GSE97050 | MAP2K3 | 5606 | A_23_P118427 | -0.1906 | 0.4518 | |
| SRP007169 | MAP2K3 | 5606 | RNAseq | -1.3897 | 0.0115 | |
| SRP008496 | MAP2K3 | 5606 | RNAseq | -1.6342 | 0.0000 | |
| SRP064894 | MAP2K3 | 5606 | RNAseq | -0.7661 | 0.0007 | |
| SRP133303 | MAP2K3 | 5606 | RNAseq | -0.4491 | 0.0006 | |
| SRP159526 | MAP2K3 | 5606 | RNAseq | -0.7004 | 0.0259 | |
| SRP193095 | MAP2K3 | 5606 | RNAseq | -0.5144 | 0.0039 | |
| SRP219564 | MAP2K3 | 5606 | RNAseq | -0.2515 | 0.4296 | |
| TCGA | MAP2K3 | 5606 | RNAseq | -0.1192 | 0.0264 |
Upregulated datasets: 0; Downregulated datasets: 2.
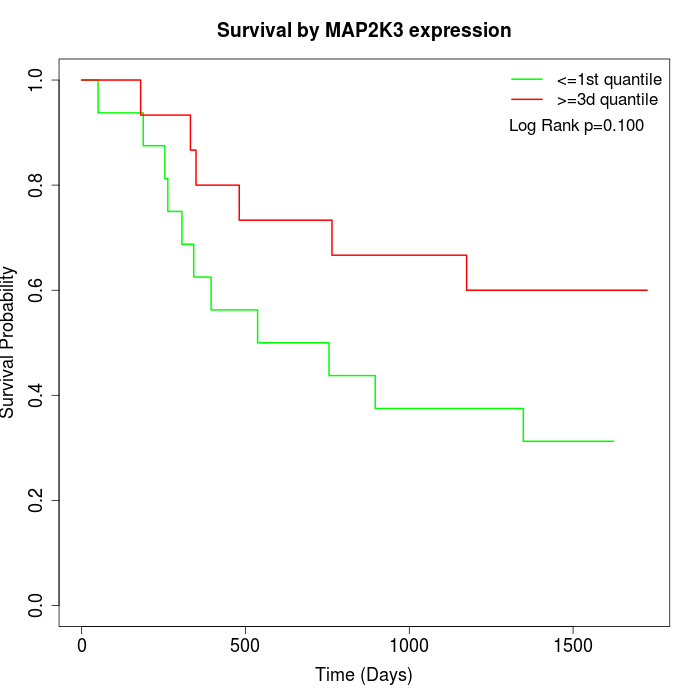
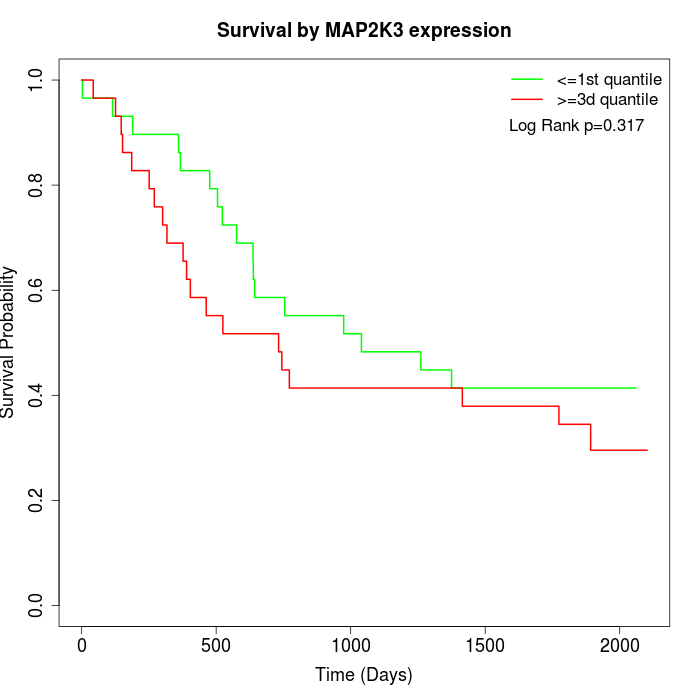
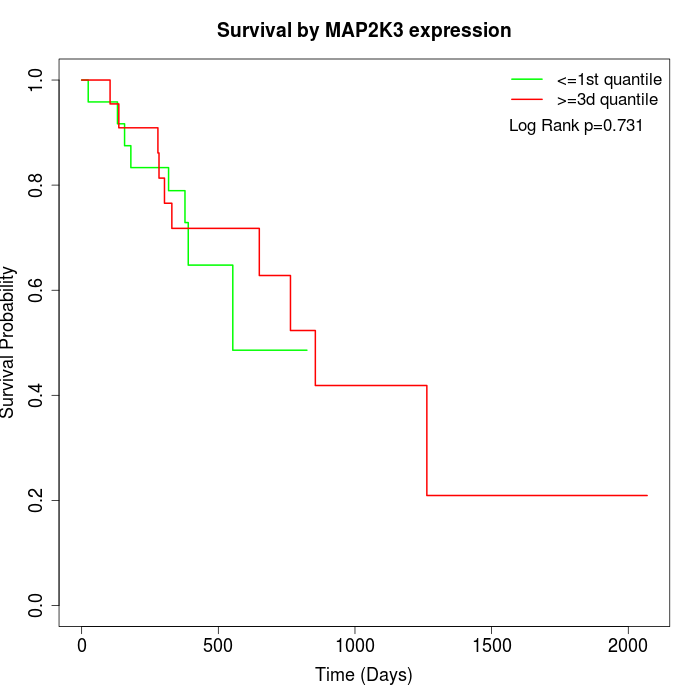
Survival by MAP2K3 expression:
Note: Click image to view full size file.
Copy number change of MAP2K3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP2K3 | 5606 | 4 | 5 | 21 | |
| GSE20123 | MAP2K3 | 5606 | 3 | 6 | 21 | |
| GSE43470 | MAP2K3 | 5606 | 1 | 5 | 37 | |
| GSE46452 | MAP2K3 | 5606 | 34 | 1 | 24 | |
| GSE47630 | MAP2K3 | 5606 | 7 | 1 | 32 | |
| GSE54993 | MAP2K3 | 5606 | 2 | 4 | 64 | |
| GSE54994 | MAP2K3 | 5606 | 6 | 6 | 41 | |
| GSE60625 | MAP2K3 | 5606 | 4 | 0 | 7 | |
| GSE74703 | MAP2K3 | 5606 | 1 | 2 | 33 | |
| GSE74704 | MAP2K3 | 5606 | 2 | 2 | 16 | |
| TCGA | MAP2K3 | 5606 | 18 | 25 | 53 |
Total number of gains: 82; Total number of losses: 57; Total Number of normals: 349.
Somatic mutations of MAP2K3:
Generating mutation plots.
Highly correlated genes for MAP2K3:
Showing top 20/113 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP2K3 | BCL10 | 0.746169 | 3 | 0 | 3 |
| MAP2K3 | LRRFIP2 | 0.746023 | 3 | 0 | 3 |
| MAP2K3 | NLRX1 | 0.739859 | 3 | 0 | 3 |
| MAP2K3 | VPS37B | 0.73753 | 3 | 0 | 3 |
| MAP2K3 | RAB5A | 0.735765 | 3 | 0 | 3 |
| MAP2K3 | COMTD1 | 0.735086 | 3 | 0 | 3 |
| MAP2K3 | PI4K2A | 0.734945 | 3 | 0 | 3 |
| MAP2K3 | ZBTB7A | 0.73389 | 3 | 0 | 3 |
| MAP2K3 | PLEKHM1 | 0.733548 | 3 | 0 | 3 |
| MAP2K3 | SERTAD1 | 0.731551 | 3 | 0 | 3 |
| MAP2K3 | CLTB | 0.728248 | 3 | 0 | 3 |
| MAP2K3 | RABGGTA | 0.725363 | 3 | 0 | 3 |
| MAP2K3 | TOM1 | 0.714827 | 3 | 0 | 3 |
| MAP2K3 | KCNG2 | 0.714351 | 3 | 0 | 3 |
| MAP2K3 | SH3GLB1 | 0.714333 | 3 | 0 | 3 |
| MAP2K3 | SNX33 | 0.709943 | 3 | 0 | 3 |
| MAP2K3 | CHMP2A | 0.706312 | 3 | 0 | 3 |
| MAP2K3 | MTF1 | 0.706141 | 3 | 0 | 3 |
| MAP2K3 | TNIP1 | 0.704899 | 3 | 0 | 3 |
| MAP2K3 | GTF3C1 | 0.701576 | 3 | 0 | 3 |
For details and further investigation, click here