| Full name: potassium voltage-gated channel modifier subfamily G member 4 | Alias Symbol: Kv6.4 | ||
| Type: protein-coding gene | Cytoband: 16q24.1 | ||
| Entrez ID: 93107 | HGNC ID: HGNC:19697 | Ensembl Gene: ENSG00000168418 | OMIM ID: 607603 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNG4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNG4 | 93107 | 1552598_at | -0.2137 | 0.5069 | |
| GSE26886 | KCNG4 | 93107 | 1552598_at | -0.0351 | 0.7810 | |
| GSE45670 | KCNG4 | 93107 | 1552598_at | -0.1041 | 0.3331 | |
| GSE53622 | KCNG4 | 93107 | 92434 | -0.0801 | 0.4115 | |
| GSE53624 | KCNG4 | 93107 | 92434 | -0.2578 | 0.0063 | |
| GSE63941 | KCNG4 | 93107 | 1552598_at | 0.0363 | 0.8264 | |
| GSE77861 | KCNG4 | 93107 | 1552598_at | -0.0108 | 0.9184 | |
| GSE97050 | KCNG4 | 93107 | A_23_P365248 | -0.0301 | 0.9469 |
Upregulated datasets: 0; Downregulated datasets: 0.
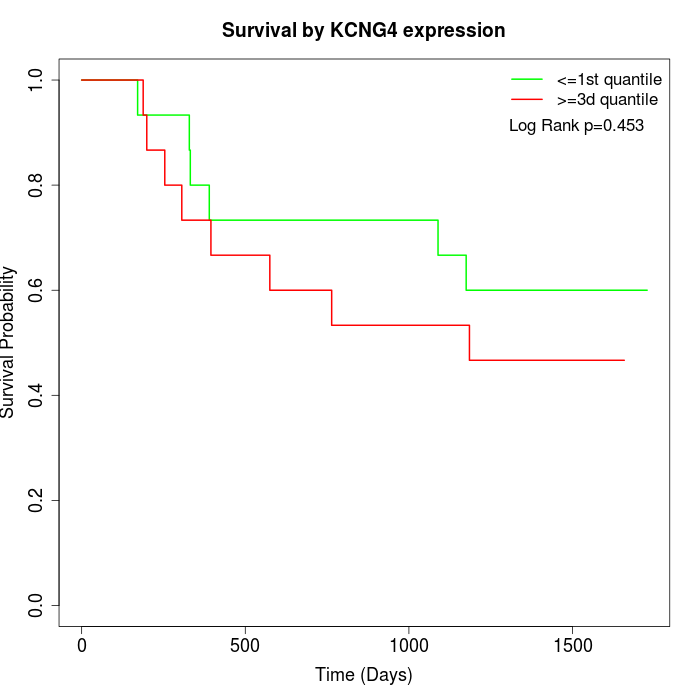
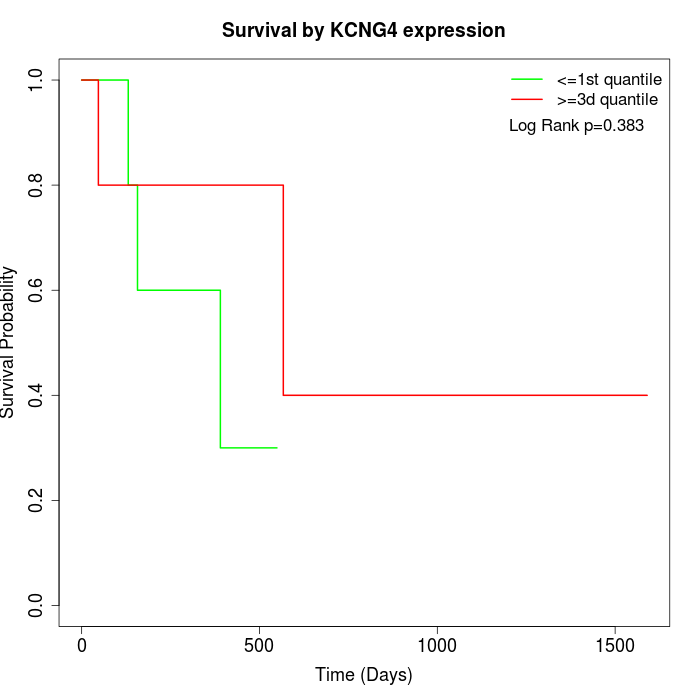
Survival by KCNG4 expression:
Note: Click image to view full size file.
Copy number change of KCNG4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNG4 | 93107 | 4 | 4 | 22 | |
| GSE20123 | KCNG4 | 93107 | 4 | 4 | 22 | |
| GSE43470 | KCNG4 | 93107 | 2 | 10 | 31 | |
| GSE46452 | KCNG4 | 93107 | 37 | 2 | 20 | |
| GSE47630 | KCNG4 | 93107 | 11 | 9 | 20 | |
| GSE54993 | KCNG4 | 93107 | 3 | 4 | 63 | |
| GSE54994 | KCNG4 | 93107 | 8 | 10 | 35 | |
| GSE60625 | KCNG4 | 93107 | 4 | 0 | 7 | |
| GSE74703 | KCNG4 | 93107 | 2 | 7 | 27 | |
| GSE74704 | KCNG4 | 93107 | 3 | 2 | 15 | |
| TCGA | KCNG4 | 93107 | 26 | 16 | 54 |
Total number of gains: 104; Total number of losses: 68; Total Number of normals: 316.
Somatic mutations of KCNG4:
Generating mutation plots.
Highly correlated genes for KCNG4:
Showing top 20/148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNG4 | C11orf42 | 0.695384 | 3 | 0 | 3 |
| KCNG4 | AKAP14 | 0.675987 | 3 | 0 | 3 |
| KCNG4 | SLC4A4 | 0.67048 | 3 | 0 | 3 |
| KCNG4 | WEE1 | 0.66598 | 3 | 0 | 3 |
| KCNG4 | CSRP3 | 0.662992 | 3 | 0 | 3 |
| KCNG4 | MPZ | 0.655293 | 3 | 0 | 3 |
| KCNG4 | GPR3 | 0.651011 | 3 | 0 | 3 |
| KCNG4 | HTR1E | 0.649726 | 3 | 0 | 3 |
| KCNG4 | PREX2 | 0.646107 | 3 | 0 | 3 |
| KCNG4 | RET | 0.640915 | 3 | 0 | 3 |
| KCNG4 | PDE5A | 0.640201 | 4 | 0 | 4 |
| KCNG4 | SRRM2-AS1 | 0.635189 | 3 | 0 | 3 |
| KCNG4 | ATRNL1 | 0.631281 | 3 | 0 | 3 |
| KCNG4 | BMX | 0.630519 | 4 | 0 | 3 |
| KCNG4 | E2F4 | 0.629339 | 5 | 0 | 5 |
| KCNG4 | TSHB | 0.628477 | 4 | 0 | 4 |
| KCNG4 | PSG1 | 0.627241 | 3 | 0 | 3 |
| KCNG4 | TNFAIP8L3 | 0.617775 | 3 | 0 | 3 |
| KCNG4 | ANHX | 0.616688 | 3 | 0 | 3 |
| KCNG4 | PRB1 | 0.616302 | 4 | 0 | 3 |
For details and further investigation, click here