| Full name: potassium inwardly rectifying channel subfamily J member 1 | Alias Symbol: Kir1.1|ROMK1 | ||
| Type: protein-coding gene | Cytoband: 11q24.3 | ||
| Entrez ID: 3758 | HGNC ID: HGNC:6255 | Ensembl Gene: ENSG00000151704 | OMIM ID: 600359 |
| Drug and gene relationship at DGIdb | |||
KCNJ1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04960 | Aldosterone-regulated sodium reabsorption | |
| hsa04971 | Gastric acid secretion |
Expression of KCNJ1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ1 | 3758 | 210402_at | -0.1042 | 0.7221 | |
| GSE20347 | KCNJ1 | 3758 | 210402_at | -0.0373 | 0.5625 | |
| GSE23400 | KCNJ1 | 3758 | 210402_at | -0.1392 | 0.0000 | |
| GSE26886 | KCNJ1 | 3758 | 210402_at | -0.0738 | 0.5383 | |
| GSE29001 | KCNJ1 | 3758 | 210403_s_at | -0.3151 | 0.0225 | |
| GSE38129 | KCNJ1 | 3758 | 210403_s_at | -0.0594 | 0.3336 | |
| GSE45670 | KCNJ1 | 3758 | 210403_s_at | -0.0037 | 0.9645 | |
| GSE53622 | KCNJ1 | 3758 | 5497 | -0.2185 | 0.1479 | |
| GSE53624 | KCNJ1 | 3758 | 5497 | -0.3177 | 0.0132 | |
| GSE63941 | KCNJ1 | 3758 | 210403_s_at | 0.1703 | 0.3344 | |
| GSE77861 | KCNJ1 | 3758 | 210403_s_at | -0.1695 | 0.0637 | |
| TCGA | KCNJ1 | 3758 | RNAseq | 0.7768 | 0.1558 |
Upregulated datasets: 0; Downregulated datasets: 0.
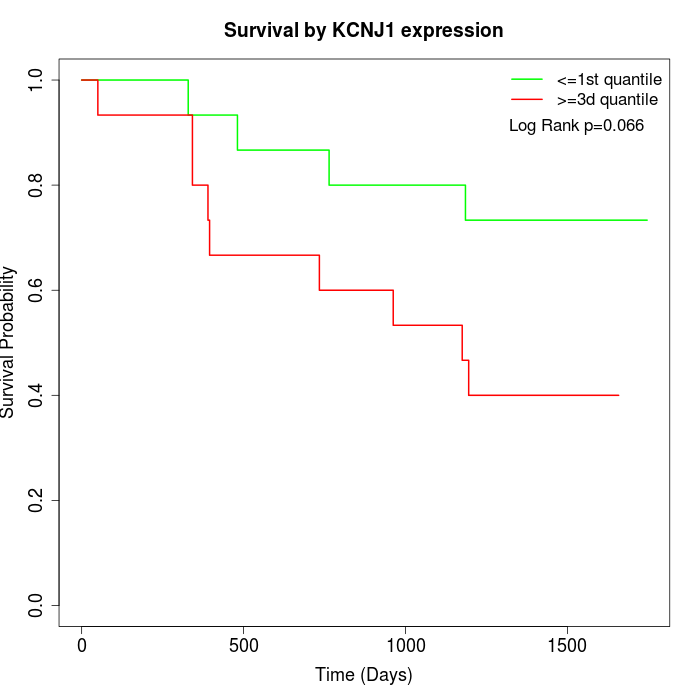
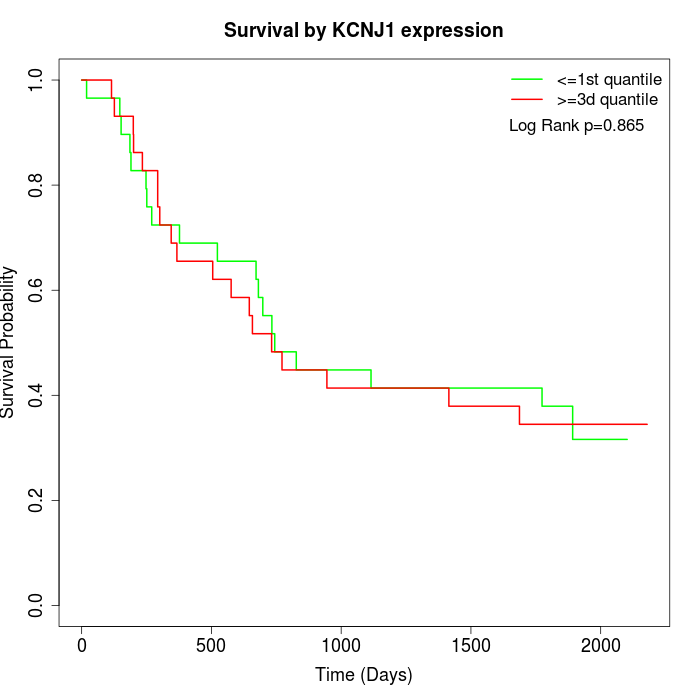
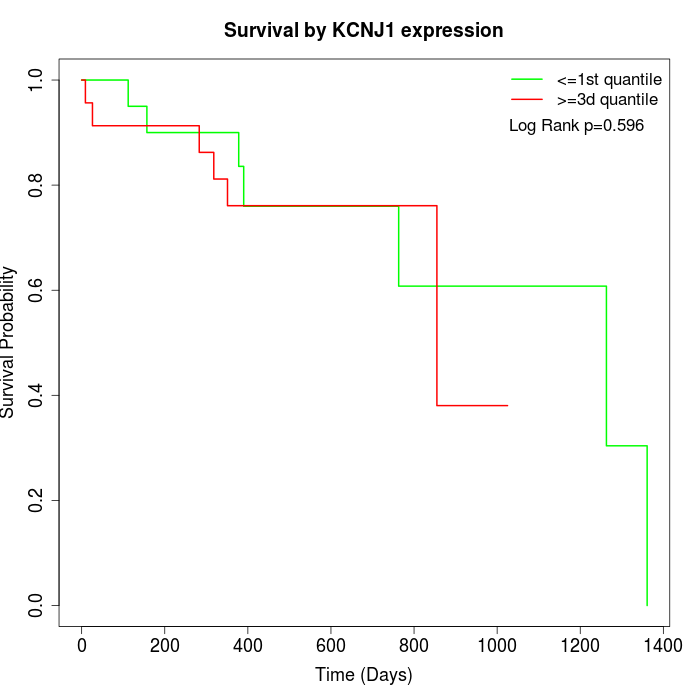
Survival by KCNJ1 expression:
Note: Click image to view full size file.
Copy number change of KCNJ1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ1 | 3758 | 0 | 13 | 17 | |
| GSE20123 | KCNJ1 | 3758 | 0 | 13 | 17 | |
| GSE43470 | KCNJ1 | 3758 | 2 | 7 | 34 | |
| GSE46452 | KCNJ1 | 3758 | 3 | 26 | 30 | |
| GSE47630 | KCNJ1 | 3758 | 2 | 19 | 19 | |
| GSE54993 | KCNJ1 | 3758 | 10 | 0 | 60 | |
| GSE54994 | KCNJ1 | 3758 | 5 | 19 | 29 | |
| GSE60625 | KCNJ1 | 3758 | 0 | 3 | 8 | |
| GSE74703 | KCNJ1 | 3758 | 1 | 5 | 30 | |
| GSE74704 | KCNJ1 | 3758 | 0 | 9 | 11 | |
| TCGA | KCNJ1 | 3758 | 6 | 50 | 40 |
Total number of gains: 29; Total number of losses: 164; Total Number of normals: 295.
Somatic mutations of KCNJ1:
Generating mutation plots.
Highly correlated genes for KCNJ1:
Showing top 20/760 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ1 | SYT13 | 0.722772 | 3 | 0 | 3 |
| KCNJ1 | HRC | 0.708494 | 3 | 0 | 3 |
| KCNJ1 | IL1RAPL2 | 0.690342 | 4 | 0 | 4 |
| KCNJ1 | CRB1 | 0.681711 | 6 | 0 | 6 |
| KCNJ1 | CER1 | 0.681222 | 5 | 0 | 5 |
| KCNJ1 | CYP4A11 | 0.680152 | 4 | 0 | 4 |
| KCNJ1 | PHLDB1 | 0.669559 | 7 | 0 | 7 |
| KCNJ1 | APBA1 | 0.666165 | 5 | 0 | 5 |
| KCNJ1 | ANKRD55 | 0.662516 | 3 | 0 | 3 |
| KCNJ1 | THRB | 0.662166 | 3 | 0 | 3 |
| KCNJ1 | SERPINA4 | 0.66189 | 4 | 0 | 4 |
| KCNJ1 | C2CD2L | 0.659981 | 3 | 0 | 3 |
| KCNJ1 | OPN4 | 0.658743 | 3 | 0 | 3 |
| KCNJ1 | CYP2A7 | 0.658607 | 5 | 0 | 4 |
| KCNJ1 | HTR3A | 0.658278 | 5 | 0 | 5 |
| KCNJ1 | TULP2 | 0.658224 | 6 | 0 | 6 |
| KCNJ1 | CPN1 | 0.65046 | 5 | 0 | 5 |
| KCNJ1 | CYP2D6 | 0.649736 | 6 | 0 | 6 |
| KCNJ1 | MUC5AC | 0.646142 | 4 | 0 | 4 |
| KCNJ1 | DRP2 | 0.644365 | 4 | 0 | 3 |
For details and further investigation, click here