| Full name: ankyrin repeat domain 55 | Alias Symbol: FLJ11795 | ||
| Type: protein-coding gene | Cytoband: 5q11.2 | ||
| Entrez ID: 79722 | HGNC ID: HGNC:25681 | Ensembl Gene: ENSG00000164512 | OMIM ID: 615189 |
| Drug and gene relationship at DGIdb | |||
Expression of ANKRD55:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ANKRD55 | 79722 | 220112_at | 0.1347 | 0.7460 | |
| GSE20347 | ANKRD55 | 79722 | 220112_at | -0.0048 | 0.9525 | |
| GSE23400 | ANKRD55 | 79722 | 220112_at | -0.0947 | 0.0027 | |
| GSE26886 | ANKRD55 | 79722 | 220112_at | -0.1353 | 0.3331 | |
| GSE29001 | ANKRD55 | 79722 | 220112_at | -0.1943 | 0.1091 | |
| GSE38129 | ANKRD55 | 79722 | 220112_at | -0.1117 | 0.0658 | |
| GSE45670 | ANKRD55 | 79722 | 220112_at | -0.0798 | 0.4514 | |
| GSE53622 | ANKRD55 | 79722 | 107603 | -0.4730 | 0.0012 | |
| GSE53624 | ANKRD55 | 79722 | 107603 | -1.1299 | 0.0000 | |
| GSE63941 | ANKRD55 | 79722 | 220112_at | 0.0873 | 0.5938 | |
| GSE77861 | ANKRD55 | 79722 | 220112_at | -0.0565 | 0.5599 | |
| TCGA | ANKRD55 | 79722 | RNAseq | -0.5780 | 0.4300 |
Upregulated datasets: 0; Downregulated datasets: 1.
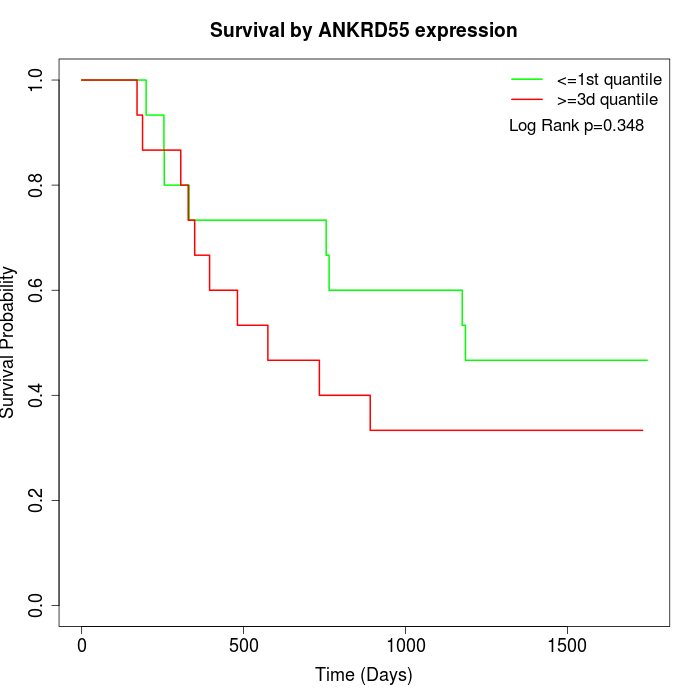
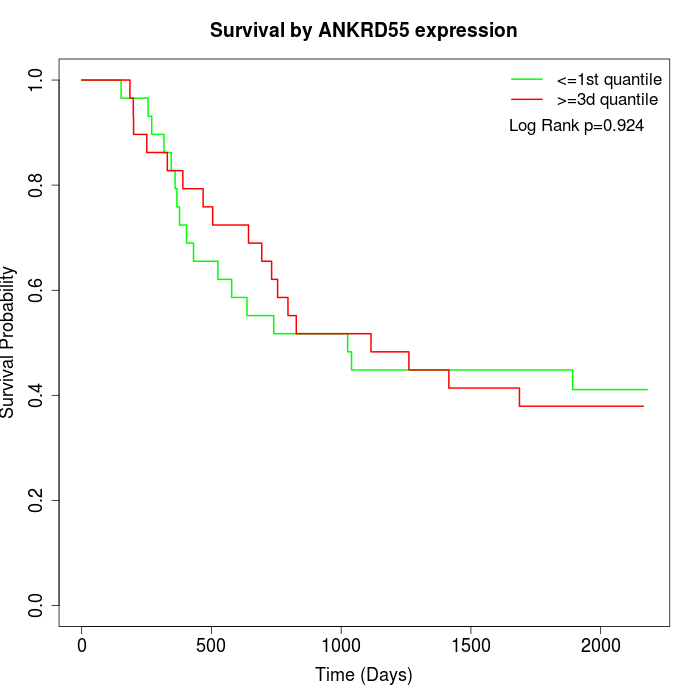
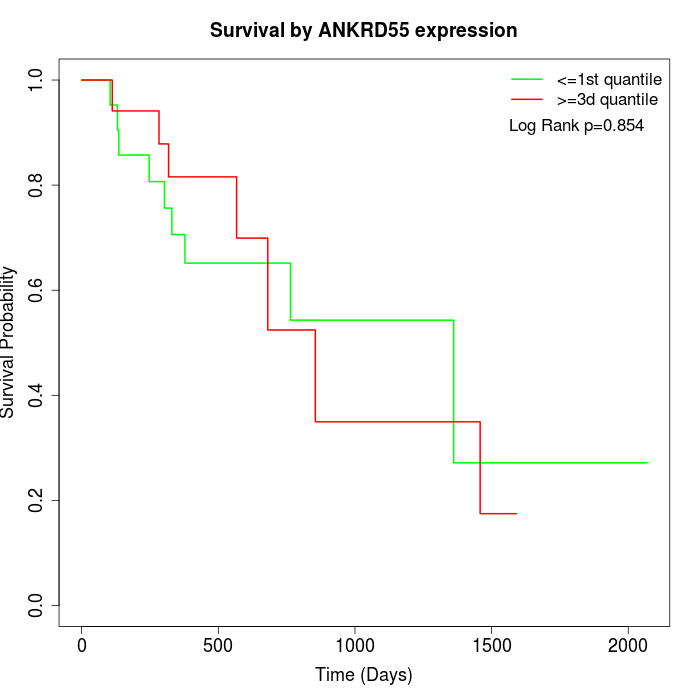
Survival by ANKRD55 expression:
Note: Click image to view full size file.
Copy number change of ANKRD55:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ANKRD55 | 79722 | 0 | 13 | 17 | |
| GSE20123 | ANKRD55 | 79722 | 0 | 13 | 17 | |
| GSE43470 | ANKRD55 | 79722 | 1 | 12 | 30 | |
| GSE46452 | ANKRD55 | 79722 | 1 | 27 | 31 | |
| GSE47630 | ANKRD55 | 79722 | 2 | 16 | 22 | |
| GSE54993 | ANKRD55 | 79722 | 8 | 1 | 61 | |
| GSE54994 | ANKRD55 | 79722 | 2 | 20 | 31 | |
| GSE60625 | ANKRD55 | 79722 | 0 | 0 | 11 | |
| GSE74703 | ANKRD55 | 79722 | 1 | 9 | 26 | |
| GSE74704 | ANKRD55 | 79722 | 0 | 6 | 14 | |
| TCGA | ANKRD55 | 79722 | 7 | 46 | 43 |
Total number of gains: 22; Total number of losses: 163; Total Number of normals: 303.
Somatic mutations of ANKRD55:
Generating mutation plots.
Highly correlated genes for ANKRD55:
Showing top 20/354 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ANKRD55 | KCNJ1 | 0.662516 | 3 | 0 | 3 |
| ANKRD55 | IFNA10 | 0.658799 | 4 | 0 | 4 |
| ANKRD55 | STRADA | 0.643539 | 3 | 0 | 3 |
| ANKRD55 | KIR3DL3 | 0.638715 | 5 | 0 | 4 |
| ANKRD55 | TNFRSF4 | 0.634109 | 3 | 0 | 3 |
| ANKRD55 | CRISP1 | 0.633944 | 5 | 0 | 4 |
| ANKRD55 | THEMIS | 0.627786 | 3 | 0 | 3 |
| ANKRD55 | OR1A1 | 0.626603 | 4 | 0 | 3 |
| ANKRD55 | OR1G1 | 0.62268 | 4 | 0 | 4 |
| ANKRD55 | GC | 0.62264 | 3 | 0 | 3 |
| ANKRD55 | PDE3B | 0.620714 | 4 | 0 | 4 |
| ANKRD55 | CD1E | 0.618723 | 4 | 0 | 3 |
| ANKRD55 | CA1 | 0.618102 | 6 | 0 | 6 |
| ANKRD55 | CYP2F1 | 0.61616 | 3 | 0 | 3 |
| ANKRD55 | SIRT3 | 0.615362 | 5 | 0 | 5 |
| ANKRD55 | SEZ6L | 0.615286 | 4 | 0 | 4 |
| ANKRD55 | EPHA5 | 0.613896 | 4 | 0 | 4 |
| ANKRD55 | TSHB | 0.61318 | 4 | 0 | 3 |
| ANKRD55 | MMP16 | 0.610366 | 3 | 0 | 3 |
| ANKRD55 | GIMAP1 | 0.609786 | 3 | 0 | 3 |
For details and further investigation, click here