| Full name: potassium inwardly rectifying channel subfamily J member 3 | Alias Symbol: Kir3.1|GIRK1|KGA | ||
| Type: protein-coding gene | Cytoband: 2q24.1 | ||
| Entrez ID: 3760 | HGNC ID: HGNC:6264 | Ensembl Gene: ENSG00000162989 | OMIM ID: 601534 |
| Drug and gene relationship at DGIdb | |||
KCNJ3 involved pathways:
Expression of KCNJ3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ3 | 3760 | 207141_s_at | -0.1377 | 0.6269 | |
| GSE20347 | KCNJ3 | 3760 | 207141_s_at | -0.0072 | 0.9501 | |
| GSE23400 | KCNJ3 | 3760 | 207141_s_at | -0.1030 | 0.0310 | |
| GSE26886 | KCNJ3 | 3760 | 207141_s_at | 0.1162 | 0.4369 | |
| GSE29001 | KCNJ3 | 3760 | 207141_s_at | -0.0770 | 0.7463 | |
| GSE38129 | KCNJ3 | 3760 | 207141_s_at | -0.1171 | 0.2465 | |
| GSE45670 | KCNJ3 | 3760 | 207141_s_at | 0.0983 | 0.3592 | |
| GSE53622 | KCNJ3 | 3760 | 37241 | -0.2045 | 0.3225 | |
| GSE53624 | KCNJ3 | 3760 | 37241 | -0.1827 | 0.1343 | |
| GSE63941 | KCNJ3 | 3760 | 207141_s_at | 0.0641 | 0.7094 | |
| GSE77861 | KCNJ3 | 3760 | 207141_s_at | -0.0415 | 0.7168 | |
| SRP133303 | KCNJ3 | 3760 | RNAseq | 0.1208 | 0.5050 | |
| SRP193095 | KCNJ3 | 3760 | RNAseq | -0.1028 | 0.7758 | |
| SRP219564 | KCNJ3 | 3760 | RNAseq | -1.0743 | 0.1956 | |
| TCGA | KCNJ3 | 3760 | RNAseq | -3.4317 | 0.0011 |
Upregulated datasets: 0; Downregulated datasets: 1.
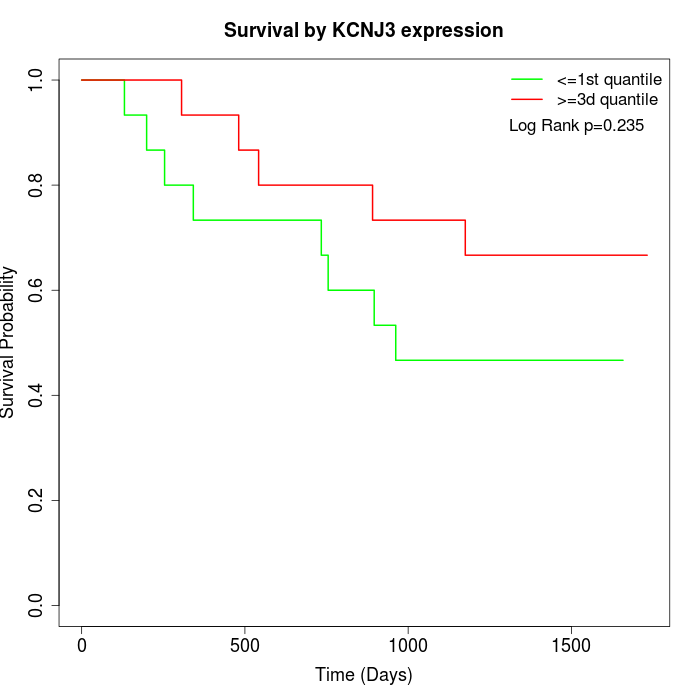
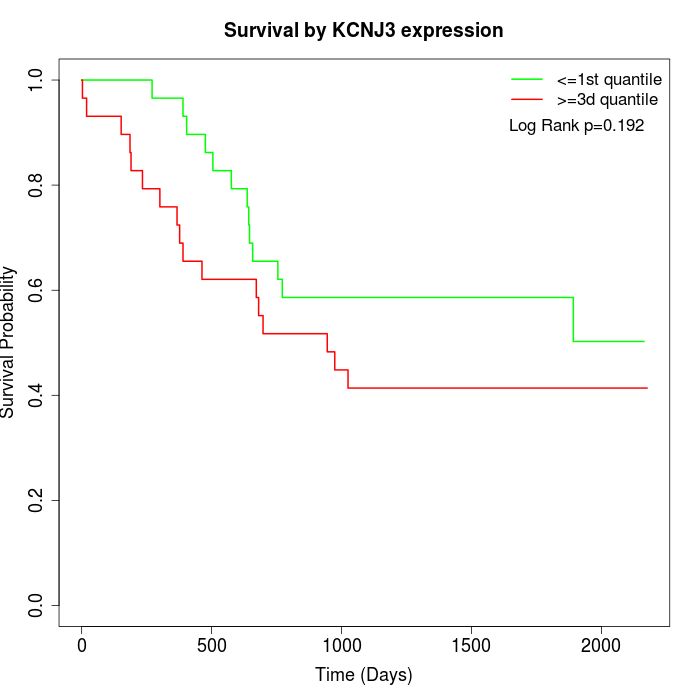
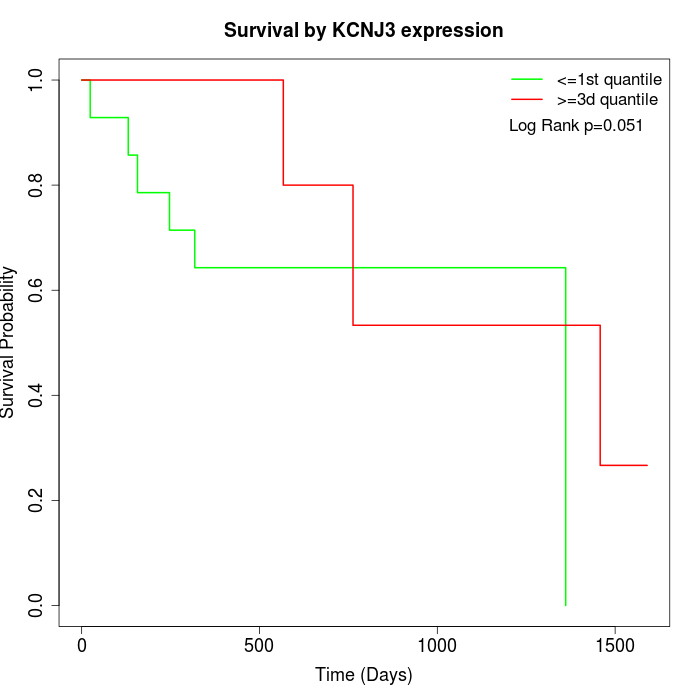
Survival by KCNJ3 expression:
Note: Click image to view full size file.
Copy number change of KCNJ3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ3 | 3760 | 4 | 2 | 24 | |
| GSE20123 | KCNJ3 | 3760 | 4 | 2 | 24 | |
| GSE43470 | KCNJ3 | 3760 | 3 | 1 | 39 | |
| GSE46452 | KCNJ3 | 3760 | 1 | 4 | 54 | |
| GSE47630 | KCNJ3 | 3760 | 5 | 3 | 32 | |
| GSE54993 | KCNJ3 | 3760 | 0 | 4 | 66 | |
| GSE54994 | KCNJ3 | 3760 | 11 | 2 | 40 | |
| GSE60625 | KCNJ3 | 3760 | 0 | 3 | 8 | |
| GSE74703 | KCNJ3 | 3760 | 2 | 1 | 33 | |
| GSE74704 | KCNJ3 | 3760 | 3 | 0 | 17 | |
| TCGA | KCNJ3 | 3760 | 21 | 9 | 66 |
Total number of gains: 54; Total number of losses: 31; Total Number of normals: 403.
Somatic mutations of KCNJ3:
Generating mutation plots.
Highly correlated genes for KCNJ3:
Showing top 20/673 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ3 | MASP2 | 0.773156 | 3 | 0 | 3 |
| KCNJ3 | CC2D2B | 0.749109 | 3 | 0 | 3 |
| KCNJ3 | ADRA1A | 0.739298 | 3 | 0 | 3 |
| KCNJ3 | HEYL | 0.716231 | 3 | 0 | 3 |
| KCNJ3 | BTNL2 | 0.714392 | 4 | 0 | 4 |
| KCNJ3 | SMCP | 0.696355 | 6 | 0 | 6 |
| KCNJ3 | IL25 | 0.694546 | 5 | 0 | 5 |
| KCNJ3 | NXPE4 | 0.692857 | 5 | 0 | 5 |
| KCNJ3 | CRTC1 | 0.691594 | 3 | 0 | 3 |
| KCNJ3 | GSX1 | 0.682302 | 3 | 0 | 3 |
| KCNJ3 | SLC5A2 | 0.679793 | 4 | 0 | 3 |
| KCNJ3 | MYO1A | 0.678922 | 3 | 0 | 3 |
| KCNJ3 | COL11A2 | 0.678801 | 4 | 0 | 4 |
| KCNJ3 | MC4R | 0.677154 | 3 | 0 | 3 |
| KCNJ3 | PRLR | 0.674284 | 4 | 0 | 3 |
| KCNJ3 | ASB12 | 0.672897 | 3 | 0 | 3 |
| KCNJ3 | MROH2A | 0.670172 | 3 | 0 | 3 |
| KCNJ3 | CYP11B2 | 0.659517 | 3 | 0 | 3 |
| KCNJ3 | EPN1 | 0.6578 | 3 | 0 | 3 |
| KCNJ3 | SLC22A13 | 0.657169 | 3 | 0 | 3 |
For details and further investigation, click here