| Full name: LSM12 homolog | Alias Symbol: FLJ30656 | ||
| Type: protein-coding gene | Cytoband: 17q21.31 | ||
| Entrez ID: 124801 | HGNC ID: HGNC:26407 | Ensembl Gene: ENSG00000161654 | OMIM ID: 611793 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of LSM12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LSM12 | 124801 | 212532_s_at | 0.0982 | 0.7875 | |
| GSE20347 | LSM12 | 124801 | 212532_s_at | -0.0551 | 0.7131 | |
| GSE23400 | LSM12 | 124801 | 212532_s_at | 0.2308 | 0.0000 | |
| GSE26886 | LSM12 | 124801 | 212532_s_at | -0.2942 | 0.0988 | |
| GSE29001 | LSM12 | 124801 | 212532_s_at | -0.0691 | 0.8919 | |
| GSE38129 | LSM12 | 124801 | 212532_s_at | 0.0862 | 0.4316 | |
| GSE45670 | LSM12 | 124801 | 212532_s_at | 0.4635 | 0.0003 | |
| GSE53622 | LSM12 | 124801 | 27140 | 0.2968 | 0.0000 | |
| GSE53624 | LSM12 | 124801 | 27140 | 0.3583 | 0.0000 | |
| GSE63941 | LSM12 | 124801 | 212532_s_at | 0.6166 | 0.0335 | |
| GSE77861 | LSM12 | 124801 | 212532_s_at | 0.2809 | 0.2737 | |
| GSE97050 | LSM12 | 124801 | A_23_P307430 | 0.2671 | 0.2850 | |
| SRP007169 | LSM12 | 124801 | RNAseq | -0.0400 | 0.8866 | |
| SRP008496 | LSM12 | 124801 | RNAseq | -0.1090 | 0.4964 | |
| SRP064894 | LSM12 | 124801 | RNAseq | 0.3071 | 0.0673 | |
| SRP133303 | LSM12 | 124801 | RNAseq | 0.4434 | 0.0012 | |
| SRP159526 | LSM12 | 124801 | RNAseq | 0.2588 | 0.1579 | |
| SRP193095 | LSM12 | 124801 | RNAseq | 0.0035 | 0.9741 | |
| SRP219564 | LSM12 | 124801 | RNAseq | 0.1413 | 0.6612 | |
| TCGA | LSM12 | 124801 | RNAseq | 0.1824 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
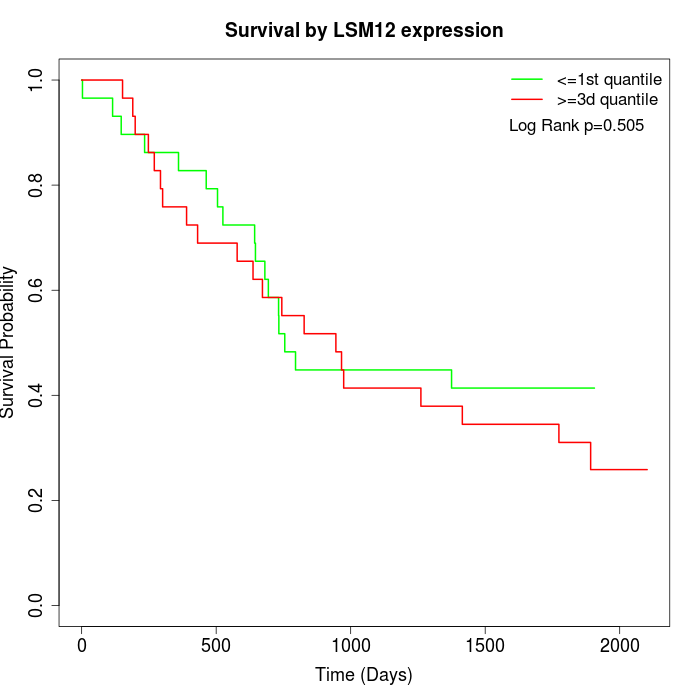
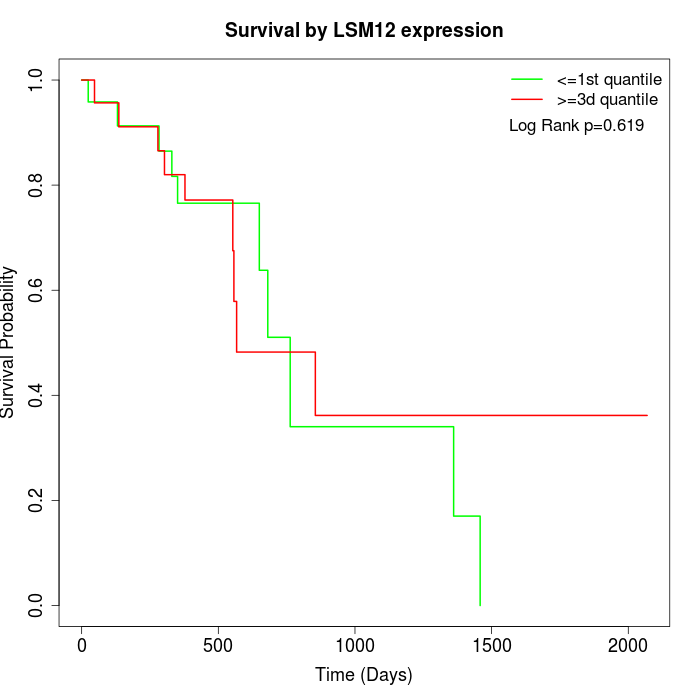
Survival by LSM12 expression:
Note: Click image to view full size file.
Copy number change of LSM12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LSM12 | 124801 | 5 | 2 | 23 | |
| GSE20123 | LSM12 | 124801 | 5 | 2 | 23 | |
| GSE43470 | LSM12 | 124801 | 1 | 2 | 40 | |
| GSE46452 | LSM12 | 124801 | 32 | 0 | 27 | |
| GSE47630 | LSM12 | 124801 | 9 | 0 | 31 | |
| GSE54993 | LSM12 | 124801 | 3 | 4 | 63 | |
| GSE54994 | LSM12 | 124801 | 8 | 6 | 39 | |
| GSE60625 | LSM12 | 124801 | 4 | 0 | 7 | |
| GSE74703 | LSM12 | 124801 | 1 | 1 | 34 | |
| GSE74704 | LSM12 | 124801 | 3 | 1 | 16 | |
| TCGA | LSM12 | 124801 | 23 | 7 | 66 |
Total number of gains: 94; Total number of losses: 25; Total Number of normals: 369.
Somatic mutations of LSM12:
Generating mutation plots.
Highly correlated genes for LSM12:
Showing top 20/726 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LSM12 | CIB1 | 0.752633 | 3 | 0 | 3 |
| LSM12 | CISD3 | 0.743715 | 3 | 0 | 3 |
| LSM12 | NOP10 | 0.743329 | 3 | 0 | 3 |
| LSM12 | LMF2 | 0.73762 | 3 | 0 | 3 |
| LSM12 | YWHAG | 0.734175 | 3 | 0 | 3 |
| LSM12 | NUDT4 | 0.722151 | 3 | 0 | 3 |
| LSM12 | RAVER1 | 0.71924 | 3 | 0 | 3 |
| LSM12 | UQCR10 | 0.712547 | 5 | 0 | 4 |
| LSM12 | NIPA2 | 0.709574 | 4 | 0 | 3 |
| LSM12 | KCNK17 | 0.709141 | 3 | 0 | 3 |
| LSM12 | AEBP2 | 0.701702 | 4 | 0 | 3 |
| LSM12 | RBFA | 0.701007 | 3 | 0 | 3 |
| LSM12 | C17orf80 | 0.690924 | 3 | 0 | 3 |
| LSM12 | DHX38 | 0.688025 | 3 | 0 | 3 |
| LSM12 | TIAL1 | 0.6821 | 3 | 0 | 3 |
| LSM12 | GHDC | 0.68111 | 3 | 0 | 3 |
| LSM12 | MYO1E | 0.680785 | 3 | 0 | 3 |
| LSM12 | KRAS | 0.679615 | 4 | 0 | 3 |
| LSM12 | FIBP | 0.678446 | 5 | 0 | 3 |
| LSM12 | DHX33 | 0.677987 | 4 | 0 | 3 |
For details and further investigation, click here