| Full name: potassium two pore domain channel subfamily K member 5 | Alias Symbol: K2p5.1|TASK-2 | ||
| Type: protein-coding gene | Cytoband: 6p21.2 | ||
| Entrez ID: 8645 | HGNC ID: HGNC:6280 | Ensembl Gene: ENSG00000164626 | OMIM ID: 603493 |
| Drug and gene relationship at DGIdb | |||
KCNK5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04742 | Taste transduction |
Expression of KCNK5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNK5 | 8645 | 219615_s_at | 0.2099 | 0.7346 | |
| GSE20347 | KCNK5 | 8645 | 219615_s_at | -0.3070 | 0.0087 | |
| GSE23400 | KCNK5 | 8645 | 219615_s_at | -0.2571 | 0.0000 | |
| GSE26886 | KCNK5 | 8645 | 219615_s_at | -0.1816 | 0.5675 | |
| GSE29001 | KCNK5 | 8645 | 219615_s_at | 0.0231 | 0.9413 | |
| GSE38129 | KCNK5 | 8645 | 219615_s_at | -0.4694 | 0.0003 | |
| GSE45670 | KCNK5 | 8645 | 219615_s_at | -0.2427 | 0.1431 | |
| GSE53622 | KCNK5 | 8645 | 110010 | -0.9564 | 0.0000 | |
| GSE53624 | KCNK5 | 8645 | 110010 | -0.8078 | 0.0000 | |
| GSE63941 | KCNK5 | 8645 | 219615_s_at | 0.5885 | 0.1706 | |
| GSE77861 | KCNK5 | 8645 | 219615_s_at | -0.3760 | 0.0135 | |
| GSE97050 | KCNK5 | 8645 | A_23_P319423 | -0.0077 | 0.9812 | |
| SRP007169 | KCNK5 | 8645 | RNAseq | -3.9762 | 0.0000 | |
| SRP064894 | KCNK5 | 8645 | RNAseq | -1.0872 | 0.0000 | |
| SRP133303 | KCNK5 | 8645 | RNAseq | -1.0823 | 0.0007 | |
| SRP159526 | KCNK5 | 8645 | RNAseq | 0.0769 | 0.9253 | |
| SRP193095 | KCNK5 | 8645 | RNAseq | -1.0604 | 0.0118 | |
| SRP219564 | KCNK5 | 8645 | RNAseq | -0.8551 | 0.2002 | |
| TCGA | KCNK5 | 8645 | RNAseq | -0.7052 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 4.
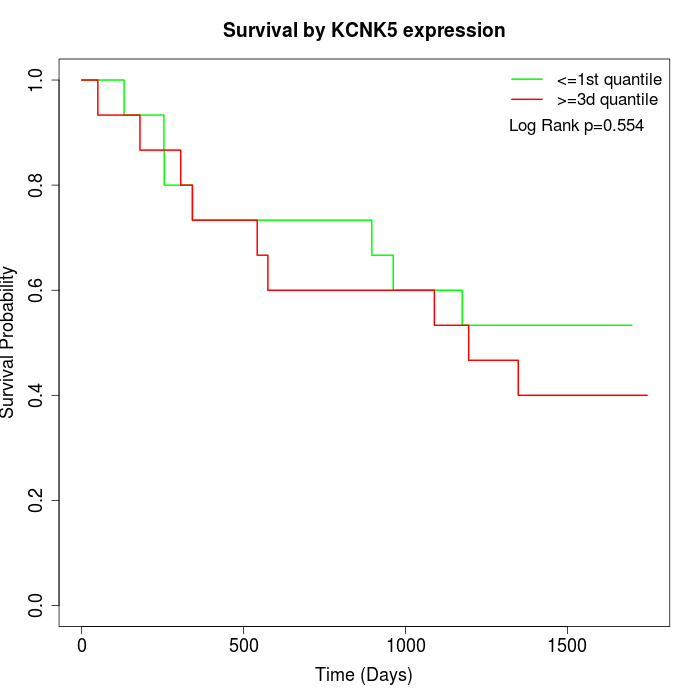
Survival by KCNK5 expression:
Note: Click image to view full size file.
Copy number change of KCNK5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNK5 | 8645 | 4 | 1 | 25 | |
| GSE20123 | KCNK5 | 8645 | 4 | 1 | 25 | |
| GSE43470 | KCNK5 | 8645 | 6 | 0 | 37 | |
| GSE46452 | KCNK5 | 8645 | 2 | 9 | 48 | |
| GSE47630 | KCNK5 | 8645 | 8 | 4 | 28 | |
| GSE54993 | KCNK5 | 8645 | 3 | 2 | 65 | |
| GSE54994 | KCNK5 | 8645 | 11 | 4 | 38 | |
| GSE60625 | KCNK5 | 8645 | 0 | 1 | 10 | |
| GSE74703 | KCNK5 | 8645 | 6 | 0 | 30 | |
| GSE74704 | KCNK5 | 8645 | 1 | 1 | 18 | |
| TCGA | KCNK5 | 8645 | 17 | 14 | 65 |
Total number of gains: 62; Total number of losses: 37; Total Number of normals: 389.
Somatic mutations of KCNK5:
Generating mutation plots.
Highly correlated genes for KCNK5:
Showing top 20/96 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNK5 | RXFP3 | 0.729186 | 3 | 0 | 3 |
| KCNK5 | LMAN1L | 0.709832 | 3 | 0 | 3 |
| KCNK5 | RNF180 | 0.698445 | 3 | 0 | 3 |
| KCNK5 | MYH13 | 0.689489 | 3 | 0 | 3 |
| KCNK5 | PCP2 | 0.668909 | 3 | 0 | 3 |
| KCNK5 | KRT35 | 0.655035 | 3 | 0 | 3 |
| KCNK5 | GALR3 | 0.65372 | 5 | 0 | 5 |
| KCNK5 | LMBR1L | 0.64662 | 3 | 0 | 3 |
| KCNK5 | SUMF1 | 0.643793 | 3 | 0 | 3 |
| KCNK5 | TMEM51-AS1 | 0.639703 | 3 | 0 | 3 |
| KCNK5 | EPHA10 | 0.639456 | 3 | 0 | 3 |
| KCNK5 | DIO2 | 0.632758 | 4 | 0 | 3 |
| KCNK5 | RETN | 0.630952 | 4 | 0 | 3 |
| KCNK5 | PROCA1 | 0.630566 | 3 | 0 | 3 |
| KCNK5 | MFNG | 0.628648 | 3 | 0 | 3 |
| KCNK5 | TNNC2 | 0.627803 | 4 | 0 | 3 |
| KCNK5 | FTCD | 0.625563 | 4 | 0 | 3 |
| KCNK5 | AZU1 | 0.622521 | 5 | 0 | 5 |
| KCNK5 | FAM110B | 0.62171 | 4 | 0 | 3 |
| KCNK5 | TCTE1 | 0.618215 | 3 | 0 | 3 |
For details and further investigation, click here