| Full name: potassium calcium-activated channel subfamily M regulatory beta subunit 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q15 | ||
| Entrez ID: 27345 | HGNC ID: HGNC:6289 | Ensembl Gene: ENSG00000135643 | OMIM ID: 605223 |
| Drug and gene relationship at DGIdb | |||
KCNMB4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04270 | Vascular smooth muscle contraction |
Expression of KCNMB4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNMB4 | 27345 | 222857_s_at | -0.1558 | 0.8865 | |
| GSE20347 | KCNMB4 | 27345 | 219287_at | 0.0175 | 0.8789 | |
| GSE23400 | KCNMB4 | 27345 | 219287_at | -0.2802 | 0.0000 | |
| GSE26886 | KCNMB4 | 27345 | 222857_s_at | 0.9166 | 0.0040 | |
| GSE29001 | KCNMB4 | 27345 | 219287_at | -0.1982 | 0.3549 | |
| GSE38129 | KCNMB4 | 27345 | 219287_at | -0.2205 | 0.1130 | |
| GSE45670 | KCNMB4 | 27345 | 222857_s_at | -0.1496 | 0.6468 | |
| GSE53622 | KCNMB4 | 27345 | 113308 | -0.4605 | 0.0266 | |
| GSE53624 | KCNMB4 | 27345 | 113308 | 0.3441 | 0.0388 | |
| GSE63941 | KCNMB4 | 27345 | 222857_s_at | 0.7000 | 0.4466 | |
| GSE77861 | KCNMB4 | 27345 | 219287_at | 0.1792 | 0.5737 | |
| GSE97050 | KCNMB4 | 27345 | A_23_P64792 | -0.5590 | 0.1552 | |
| SRP007169 | KCNMB4 | 27345 | RNAseq | 1.6675 | 0.0990 | |
| SRP064894 | KCNMB4 | 27345 | RNAseq | 0.6892 | 0.0487 | |
| SRP133303 | KCNMB4 | 27345 | RNAseq | -0.3363 | 0.2012 | |
| SRP159526 | KCNMB4 | 27345 | RNAseq | 0.8614 | 0.1954 | |
| SRP193095 | KCNMB4 | 27345 | RNAseq | 0.6289 | 0.0003 | |
| SRP219564 | KCNMB4 | 27345 | RNAseq | 0.0157 | 0.9872 | |
| TCGA | KCNMB4 | 27345 | RNAseq | 0.0427 | 0.8608 |
Upregulated datasets: 0; Downregulated datasets: 0.
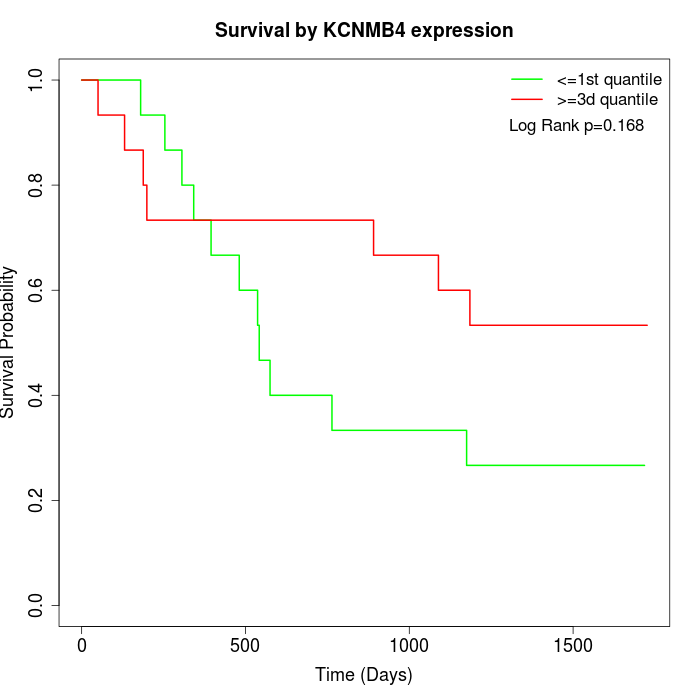
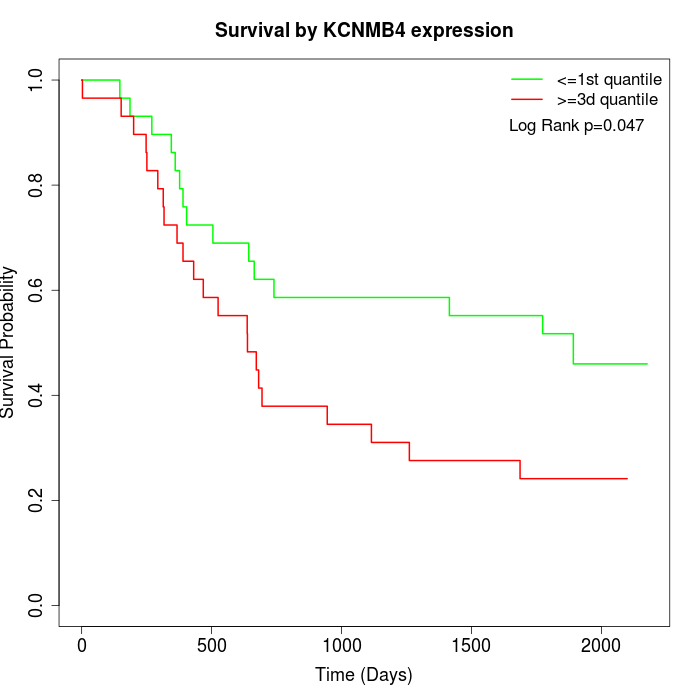
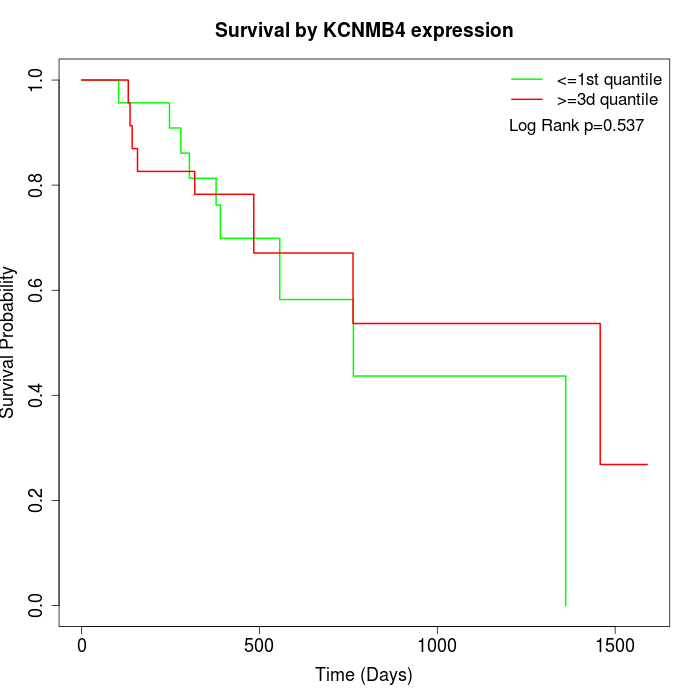
Survival by KCNMB4 expression:
Note: Click image to view full size file.
Copy number change of KCNMB4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNMB4 | 27345 | 6 | 0 | 24 | |
| GSE20123 | KCNMB4 | 27345 | 6 | 0 | 24 | |
| GSE43470 | KCNMB4 | 27345 | 4 | 0 | 39 | |
| GSE46452 | KCNMB4 | 27345 | 9 | 1 | 49 | |
| GSE47630 | KCNMB4 | 27345 | 10 | 1 | 29 | |
| GSE54993 | KCNMB4 | 27345 | 0 | 6 | 64 | |
| GSE54994 | KCNMB4 | 27345 | 8 | 1 | 44 | |
| GSE60625 | KCNMB4 | 27345 | 0 | 0 | 11 | |
| GSE74703 | KCNMB4 | 27345 | 4 | 0 | 32 | |
| GSE74704 | KCNMB4 | 27345 | 5 | 0 | 15 | |
| TCGA | KCNMB4 | 27345 | 22 | 8 | 66 |
Total number of gains: 74; Total number of losses: 17; Total Number of normals: 397.
Somatic mutations of KCNMB4:
Generating mutation plots.
Highly correlated genes for KCNMB4:
Showing top 20/704 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNMB4 | CLUAP1 | 0.792411 | 3 | 0 | 3 |
| KCNMB4 | DROSHA | 0.764001 | 3 | 0 | 3 |
| KCNMB4 | FOXC2 | 0.761532 | 3 | 0 | 3 |
| KCNMB4 | FAM124A | 0.755544 | 4 | 0 | 4 |
| KCNMB4 | SORCS1 | 0.754134 | 3 | 0 | 3 |
| KCNMB4 | TRIM47 | 0.746359 | 3 | 0 | 3 |
| KCNMB4 | LNP1 | 0.744445 | 4 | 0 | 3 |
| KCNMB4 | CYS1 | 0.743941 | 5 | 0 | 5 |
| KCNMB4 | MCM3AP | 0.743212 | 4 | 0 | 4 |
| KCNMB4 | AGBL5 | 0.741983 | 3 | 0 | 3 |
| KCNMB4 | KIAA1324L | 0.740653 | 4 | 0 | 4 |
| KCNMB4 | ZNF827 | 0.736981 | 3 | 0 | 3 |
| KCNMB4 | NINJ1 | 0.731731 | 4 | 0 | 3 |
| KCNMB4 | HCFC1 | 0.731033 | 3 | 0 | 3 |
| KCNMB4 | FAM86C1 | 0.729265 | 3 | 0 | 3 |
| KCNMB4 | SCAI | 0.727422 | 4 | 0 | 3 |
| KCNMB4 | S1PR3 | 0.71805 | 4 | 0 | 4 |
| KCNMB4 | NAA11 | 0.7157 | 3 | 0 | 3 |
| KCNMB4 | RSPO3 | 0.7144 | 5 | 0 | 5 |
| KCNMB4 | CYP2U1 | 0.713733 | 6 | 0 | 5 |
For details and further investigation, click here