| Full name: potassium voltage-gated channel subfamily Q member 2 | Alias Symbol: Kv7.2|ENB1|BFNC|KCNA11|HNSPC | ||
| Type: protein-coding gene | Cytoband: 20q13.33 | ||
| Entrez ID: 3785 | HGNC ID: HGNC:6296 | Ensembl Gene: ENSG00000075043 | OMIM ID: 602235 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNQ2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNQ2 | 3785 | 211486_s_at | -0.1655 | 0.6008 | |
| GSE20347 | KCNQ2 | 3785 | 210508_s_at | -0.2296 | 0.0004 | |
| GSE23400 | KCNQ2 | 3785 | 211486_s_at | -0.1787 | 0.0000 | |
| GSE26886 | KCNQ2 | 3785 | 211486_s_at | 0.3096 | 0.0138 | |
| GSE29001 | KCNQ2 | 3785 | 211486_s_at | -0.0506 | 0.7627 | |
| GSE38129 | KCNQ2 | 3785 | 210508_s_at | -0.2323 | 0.0155 | |
| GSE45670 | KCNQ2 | 3785 | 211486_s_at | -0.0072 | 0.9585 | |
| GSE53622 | KCNQ2 | 3785 | 45388 | -0.5001 | 0.0000 | |
| GSE53624 | KCNQ2 | 3785 | 45388 | -0.3865 | 0.0000 | |
| GSE63941 | KCNQ2 | 3785 | 211486_s_at | 0.0070 | 0.9781 | |
| GSE77861 | KCNQ2 | 3785 | 211486_s_at | -0.2043 | 0.3692 | |
| GSE97050 | KCNQ2 | 3785 | A_33_P3360758 | -0.5025 | 0.5369 | |
| TCGA | KCNQ2 | 3785 | RNAseq | -2.3623 | 0.0619 |
Upregulated datasets: 0; Downregulated datasets: 0.
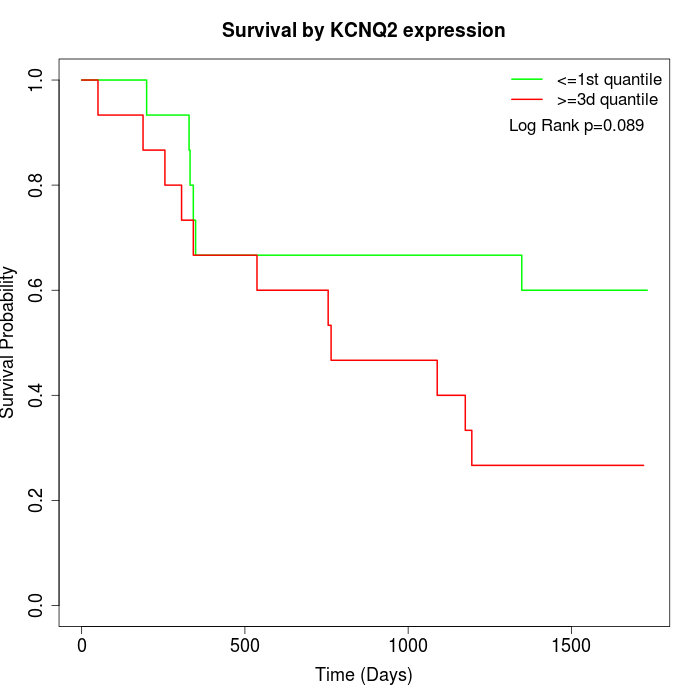
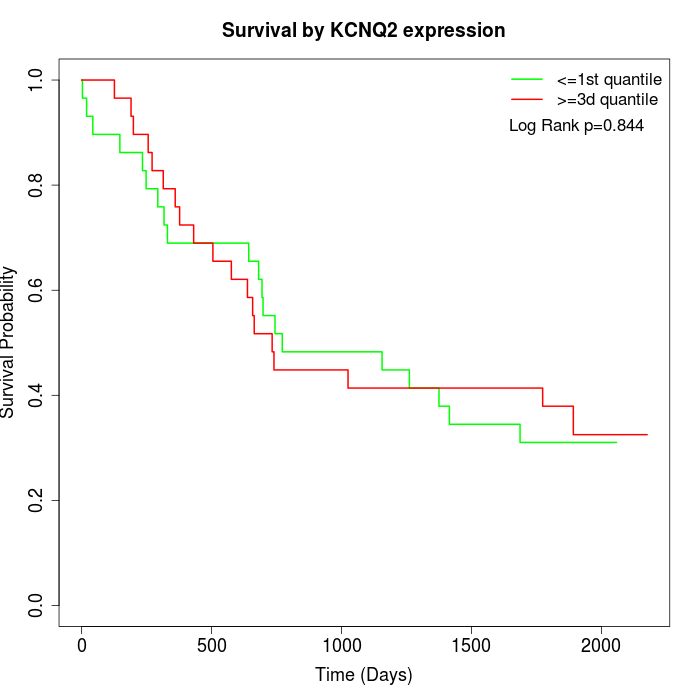
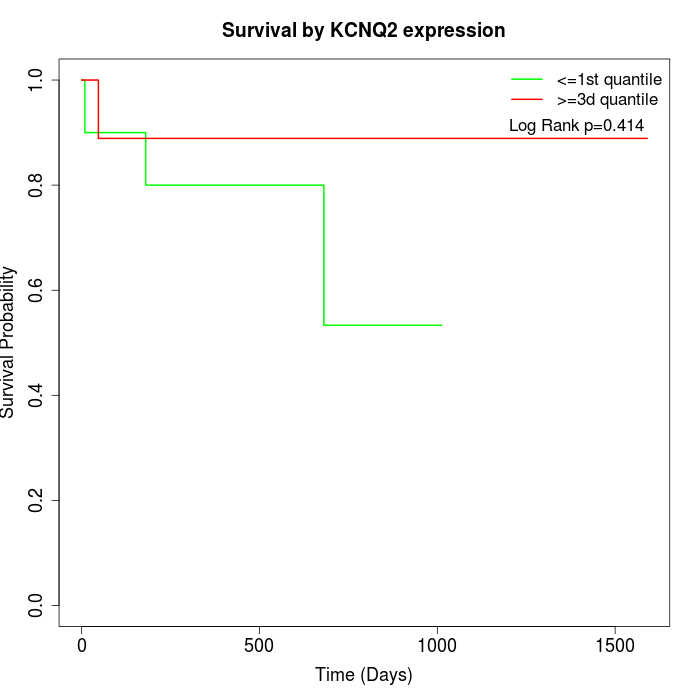
Survival by KCNQ2 expression:
Note: Click image to view full size file.
Copy number change of KCNQ2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNQ2 | 3785 | 14 | 2 | 14 | |
| GSE20123 | KCNQ2 | 3785 | 14 | 2 | 14 | |
| GSE43470 | KCNQ2 | 3785 | 11 | 1 | 31 | |
| GSE46452 | KCNQ2 | 3785 | 35 | 0 | 24 | |
| GSE47630 | KCNQ2 | 3785 | 25 | 1 | 14 | |
| GSE54993 | KCNQ2 | 3785 | 0 | 17 | 53 | |
| GSE54994 | KCNQ2 | 3785 | 29 | 0 | 24 | |
| GSE60625 | KCNQ2 | 3785 | 0 | 0 | 11 | |
| GSE74703 | KCNQ2 | 3785 | 10 | 0 | 26 | |
| GSE74704 | KCNQ2 | 3785 | 11 | 0 | 9 | |
| TCGA | KCNQ2 | 3785 | 44 | 4 | 48 |
Total number of gains: 193; Total number of losses: 27; Total Number of normals: 268.
Somatic mutations of KCNQ2:
Generating mutation plots.
Highly correlated genes for KCNQ2:
Showing top 20/668 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNQ2 | C1orf159 | 0.756732 | 3 | 0 | 3 |
| KCNQ2 | TEAD3 | 0.745316 | 4 | 0 | 4 |
| KCNQ2 | FOXP4 | 0.731231 | 3 | 0 | 3 |
| KCNQ2 | CDH15 | 0.716143 | 5 | 0 | 5 |
| KCNQ2 | YPEL2 | 0.712757 | 3 | 0 | 3 |
| KCNQ2 | OCM2 | 0.709264 | 5 | 0 | 4 |
| KCNQ2 | NPFFR1 | 0.705467 | 5 | 0 | 4 |
| KCNQ2 | CLEC14A | 0.699964 | 3 | 0 | 3 |
| KCNQ2 | FOXH1 | 0.690957 | 3 | 0 | 3 |
| KCNQ2 | ADAMTSL5 | 0.689398 | 3 | 0 | 3 |
| KCNQ2 | UNC5C | 0.689336 | 3 | 0 | 3 |
| KCNQ2 | SYN3 | 0.687812 | 4 | 0 | 4 |
| KCNQ2 | ZNF324B | 0.687483 | 3 | 0 | 3 |
| KCNQ2 | LIPF | 0.685941 | 4 | 0 | 4 |
| KCNQ2 | TMEM8B | 0.685141 | 3 | 0 | 3 |
| KCNQ2 | SLK | 0.681061 | 5 | 0 | 5 |
| KCNQ2 | HMCN2 | 0.679946 | 3 | 0 | 3 |
| KCNQ2 | ITIH1 | 0.678993 | 6 | 0 | 5 |
| KCNQ2 | USP19 | 0.675948 | 4 | 0 | 3 |
| KCNQ2 | HIC2 | 0.67498 | 4 | 0 | 4 |
For details and further investigation, click here