| Full name: potassium channel tetramerization domain containing 2 | Alias Symbol: KIAA0176 | ||
| Type: protein-coding gene | Cytoband: 17q25.1 | ||
| Entrez ID: 23510 | HGNC ID: HGNC:21294 | Ensembl Gene: ENSG00000180901 | OMIM ID: 613422 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD2 | 23510 | 34858_at | 0.1964 | 0.2672 | |
| GSE20347 | KCTD2 | 23510 | 212564_at | -0.0099 | 0.9124 | |
| GSE23400 | KCTD2 | 23510 | 34858_at | -0.0869 | 0.0736 | |
| GSE26886 | KCTD2 | 23510 | 34858_at | -0.1542 | 0.3904 | |
| GSE29001 | KCTD2 | 23510 | 34858_at | -0.0707 | 0.7151 | |
| GSE38129 | KCTD2 | 23510 | 212564_at | -0.1606 | 0.0416 | |
| GSE45670 | KCTD2 | 23510 | 34858_at | -0.0149 | 0.8921 | |
| GSE53622 | KCTD2 | 23510 | 64714 | 0.0519 | 0.3633 | |
| GSE53624 | KCTD2 | 23510 | 56924 | 0.0139 | 0.8544 | |
| GSE63941 | KCTD2 | 23510 | 34858_at | -0.2019 | 0.2743 | |
| GSE77861 | KCTD2 | 23510 | 34858_at | -0.0085 | 0.9651 | |
| GSE97050 | KCTD2 | 23510 | A_24_P324011 | -0.1294 | 0.6191 | |
| SRP007169 | KCTD2 | 23510 | RNAseq | -0.5407 | 0.0714 | |
| SRP008496 | KCTD2 | 23510 | RNAseq | -0.5009 | 0.0150 | |
| SRP064894 | KCTD2 | 23510 | RNAseq | 0.3381 | 0.0890 | |
| SRP133303 | KCTD2 | 23510 | RNAseq | 0.2073 | 0.3629 | |
| SRP159526 | KCTD2 | 23510 | RNAseq | -0.1807 | 0.3514 | |
| SRP193095 | KCTD2 | 23510 | RNAseq | 0.0421 | 0.6776 | |
| SRP219564 | KCTD2 | 23510 | RNAseq | 0.2865 | 0.4796 | |
| TCGA | KCTD2 | 23510 | RNAseq | -0.1101 | 0.0221 |
Upregulated datasets: 0; Downregulated datasets: 0.
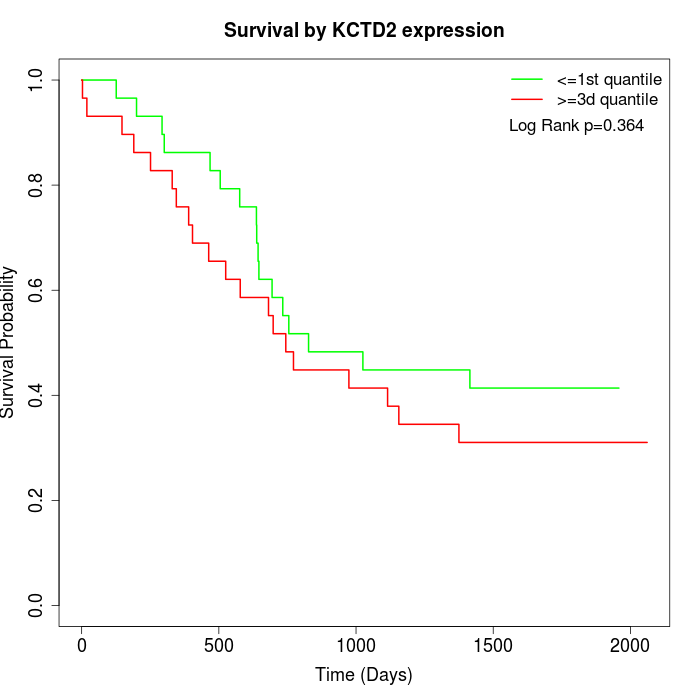
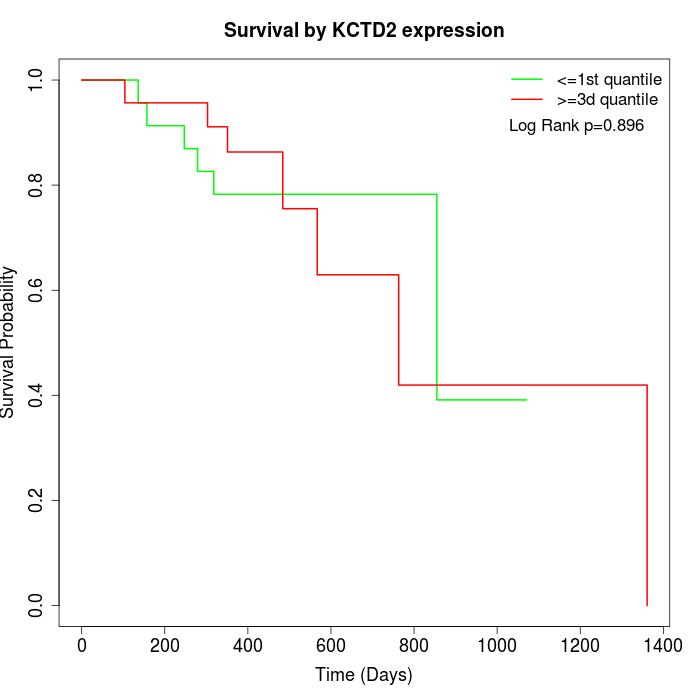
Survival by KCTD2 expression:
Note: Click image to view full size file.
Copy number change of KCTD2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD2 | 23510 | 6 | 2 | 22 | |
| GSE20123 | KCTD2 | 23510 | 6 | 2 | 22 | |
| GSE43470 | KCTD2 | 23510 | 5 | 3 | 35 | |
| GSE46452 | KCTD2 | 23510 | 33 | 0 | 26 | |
| GSE47630 | KCTD2 | 23510 | 8 | 1 | 31 | |
| GSE54993 | KCTD2 | 23510 | 2 | 5 | 63 | |
| GSE54994 | KCTD2 | 23510 | 10 | 4 | 39 | |
| GSE60625 | KCTD2 | 23510 | 6 | 0 | 5 | |
| GSE74703 | KCTD2 | 23510 | 5 | 1 | 30 | |
| GSE74704 | KCTD2 | 23510 | 4 | 1 | 15 | |
| TCGA | KCTD2 | 23510 | 32 | 10 | 54 |
Total number of gains: 117; Total number of losses: 29; Total Number of normals: 342.
Somatic mutations of KCTD2:
Generating mutation plots.
Highly correlated genes for KCTD2:
Showing top 20/163 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD2 | MOB3C | 0.798592 | 3 | 0 | 3 |
| KCTD2 | DEXI | 0.784062 | 3 | 0 | 3 |
| KCTD2 | ZACN | 0.768836 | 3 | 0 | 3 |
| KCTD2 | CHCHD6 | 0.765529 | 3 | 0 | 3 |
| KCTD2 | XPO7 | 0.753272 | 3 | 0 | 3 |
| KCTD2 | NUDT4 | 0.751988 | 3 | 0 | 3 |
| KCTD2 | BICD2 | 0.75104 | 3 | 0 | 3 |
| KCTD2 | COMMD7 | 0.737748 | 3 | 0 | 3 |
| KCTD2 | TSPAN15 | 0.736638 | 3 | 0 | 3 |
| KCTD2 | ZNF594 | 0.727261 | 3 | 0 | 3 |
| KCTD2 | KCTD10 | 0.727166 | 3 | 0 | 3 |
| KCTD2 | NSD1 | 0.715736 | 4 | 0 | 3 |
| KCTD2 | ALDOA | 0.709741 | 3 | 0 | 3 |
| KCTD2 | CTXN1 | 0.706115 | 3 | 0 | 3 |
| KCTD2 | CCDC97 | 0.700619 | 3 | 0 | 3 |
| KCTD2 | CYB5D1 | 0.699951 | 3 | 0 | 3 |
| KCTD2 | PREB | 0.698385 | 3 | 0 | 3 |
| KCTD2 | ARHGAP17 | 0.694545 | 4 | 0 | 4 |
| KCTD2 | ECSIT | 0.689693 | 3 | 0 | 3 |
| KCTD2 | GCDH | 0.68589 | 3 | 0 | 3 |
For details and further investigation, click here