| Full name: zinc activated ion channel | Alias Symbol: LGICZ|L2|ZAC|ZAC1 | ||
| Type: protein-coding gene | Cytoband: 17q25.1 | ||
| Entrez ID: 353174 | HGNC ID: HGNC:29504 | Ensembl Gene: ENSG00000186919 | OMIM ID: 610935 |
| Drug and gene relationship at DGIdb | |||
Expression of ZACN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZACN | 353174 | 1555550_at | 0.0435 | 0.8659 | |
| GSE26886 | ZACN | 353174 | 1555550_at | -0.2106 | 0.0543 | |
| GSE45670 | ZACN | 353174 | 1555550_at | 0.0947 | 0.3156 | |
| GSE53622 | ZACN | 353174 | 50902 | -0.4177 | 0.0000 | |
| GSE53624 | ZACN | 353174 | 50902 | -0.2362 | 0.0001 | |
| GSE63941 | ZACN | 353174 | 1555550_at | -0.1807 | 0.1933 | |
| GSE77861 | ZACN | 353174 | 1555550_at | -0.2516 | 0.1114 | |
| GSE97050 | ZACN | 353174 | A_23_P316064 | -0.3517 | 0.2433 | |
| SRP133303 | ZACN | 353174 | RNAseq | -0.6092 | 0.2581 | |
| TCGA | ZACN | 353174 | RNAseq | 0.6789 | 0.1597 |
Upregulated datasets: 0; Downregulated datasets: 0.
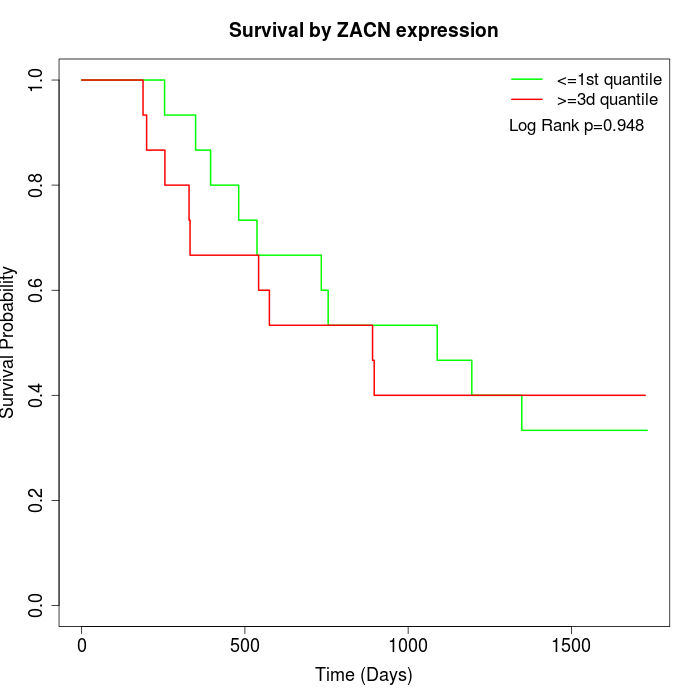
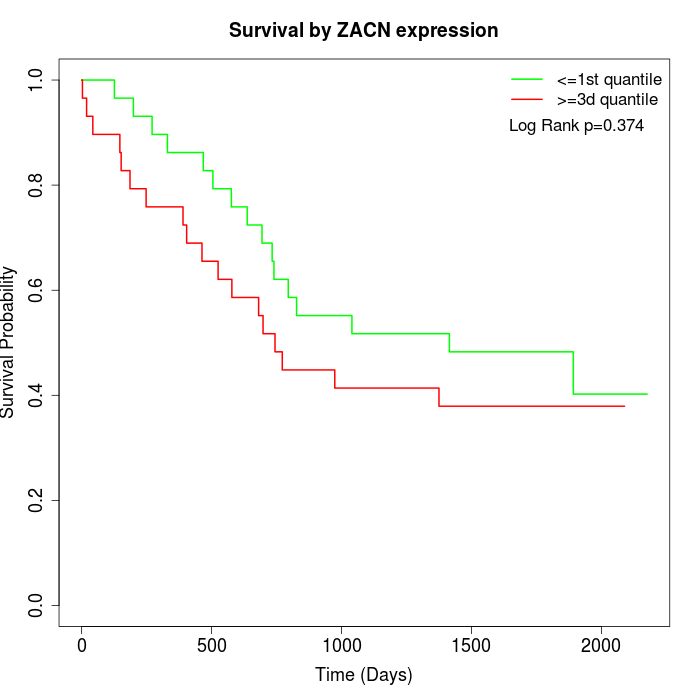
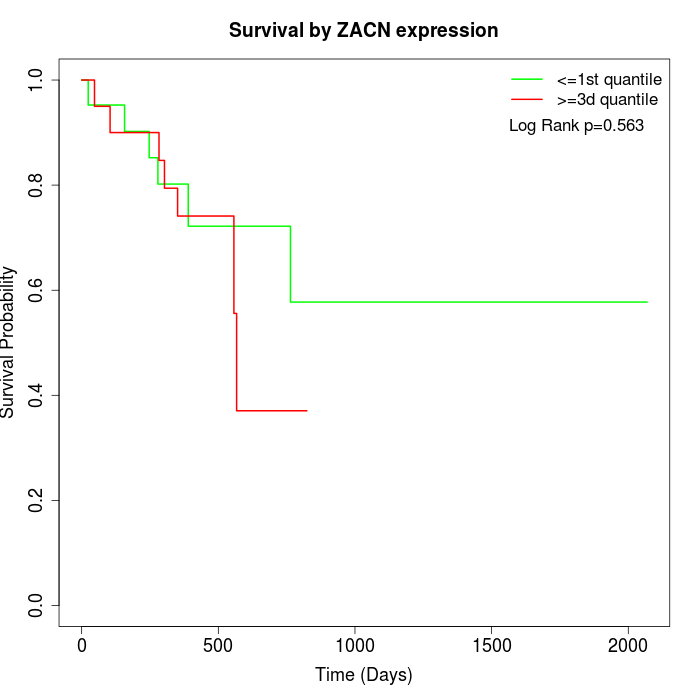
Survival by ZACN expression:
Note: Click image to view full size file.
Copy number change of ZACN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZACN | 353174 | 7 | 2 | 21 | |
| GSE20123 | ZACN | 353174 | 7 | 2 | 21 | |
| GSE43470 | ZACN | 353174 | 5 | 3 | 35 | |
| GSE46452 | ZACN | 353174 | 34 | 0 | 25 | |
| GSE47630 | ZACN | 353174 | 8 | 1 | 31 | |
| GSE54993 | ZACN | 353174 | 2 | 5 | 63 | |
| GSE54994 | ZACN | 353174 | 10 | 4 | 39 | |
| GSE60625 | ZACN | 353174 | 6 | 0 | 5 | |
| GSE74703 | ZACN | 353174 | 5 | 1 | 30 | |
| GSE74704 | ZACN | 353174 | 5 | 1 | 14 | |
| TCGA | ZACN | 353174 | 31 | 10 | 55 |
Total number of gains: 120; Total number of losses: 29; Total Number of normals: 339.
Somatic mutations of ZACN:
Generating mutation plots.
Highly correlated genes for ZACN:
Showing top 20/375 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZACN | MRPS18A | 0.77838 | 3 | 0 | 3 |
| ZACN | KCTD2 | 0.768836 | 3 | 0 | 3 |
| ZACN | PARD3 | 0.765991 | 3 | 0 | 3 |
| ZACN | USO1 | 0.761817 | 3 | 0 | 3 |
| ZACN | FKBP8 | 0.754001 | 3 | 0 | 3 |
| ZACN | CDKL1 | 0.746723 | 3 | 0 | 3 |
| ZACN | LMO2 | 0.741068 | 3 | 0 | 3 |
| ZACN | HSD17B10 | 0.735402 | 4 | 0 | 4 |
| ZACN | BIRC6 | 0.730777 | 3 | 0 | 3 |
| ZACN | LMO4 | 0.730651 | 3 | 0 | 3 |
| ZACN | GNPTAB | 0.728451 | 3 | 0 | 3 |
| ZACN | NUDT4 | 0.725824 | 3 | 0 | 3 |
| ZACN | DMXL1 | 0.723674 | 3 | 0 | 3 |
| ZACN | MCCC1 | 0.721946 | 3 | 0 | 3 |
| ZACN | TRIM33 | 0.721878 | 3 | 0 | 3 |
| ZACN | MORN2 | 0.719795 | 3 | 0 | 3 |
| ZACN | NANS | 0.718976 | 3 | 0 | 3 |
| ZACN | PDIK1L | 0.716016 | 3 | 0 | 3 |
| ZACN | TMEM53 | 0.715723 | 3 | 0 | 3 |
| ZACN | ZCCHC10 | 0.715578 | 3 | 0 | 3 |
For details and further investigation, click here