| Full name: cortexin 1 | Alias Symbol: FLJ25968 | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 404217 | HGNC ID: HGNC:31108 | Ensembl Gene: ENSG00000178531 | OMIM ID: 600135 |
| Drug and gene relationship at DGIdb | |||
Expression of CTXN1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTXN1 | 404217 | 228126_x_at | 0.2096 | 0.3472 | |
| GSE26886 | CTXN1 | 404217 | 228126_x_at | 0.0757 | 0.6116 | |
| GSE45670 | CTXN1 | 404217 | 228126_x_at | 0.0927 | 0.3581 | |
| GSE53622 | CTXN1 | 404217 | 83796 | 0.2845 | 0.0155 | |
| GSE53624 | CTXN1 | 404217 | 83796 | 0.4415 | 0.0000 | |
| GSE63941 | CTXN1 | 404217 | 228126_x_at | 0.3833 | 0.2033 | |
| GSE77861 | CTXN1 | 404217 | 228126_x_at | 0.0521 | 0.6853 | |
| GSE97050 | CTXN1 | 404217 | A_33_P3320953 | -0.3045 | 0.4224 | |
| SRP007169 | CTXN1 | 404217 | RNAseq | 2.5223 | 0.0255 | |
| SRP064894 | CTXN1 | 404217 | RNAseq | 1.0070 | 0.0412 | |
| SRP133303 | CTXN1 | 404217 | RNAseq | 0.8648 | 0.0672 | |
| SRP159526 | CTXN1 | 404217 | RNAseq | 1.0525 | 0.1430 | |
| SRP219564 | CTXN1 | 404217 | RNAseq | -1.0382 | 0.3570 | |
| TCGA | CTXN1 | 404217 | RNAseq | 0.2256 | 0.2494 |
Upregulated datasets: 2; Downregulated datasets: 0.
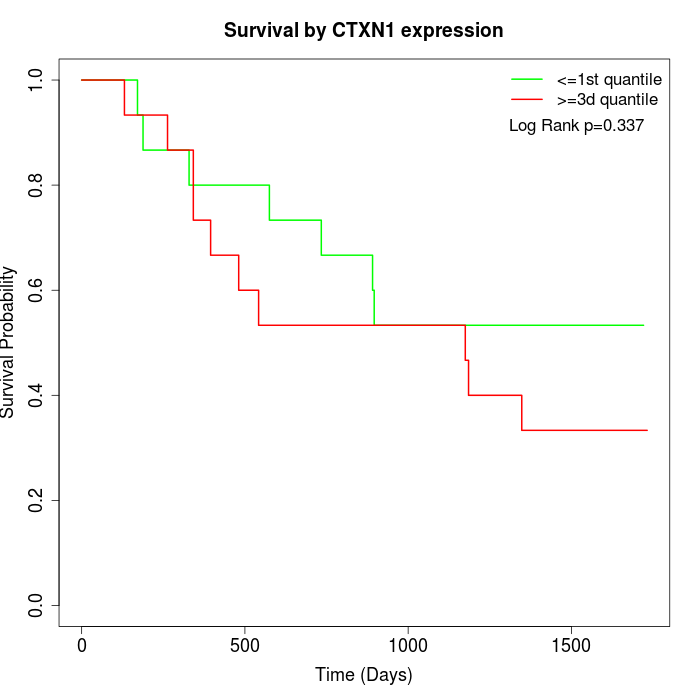
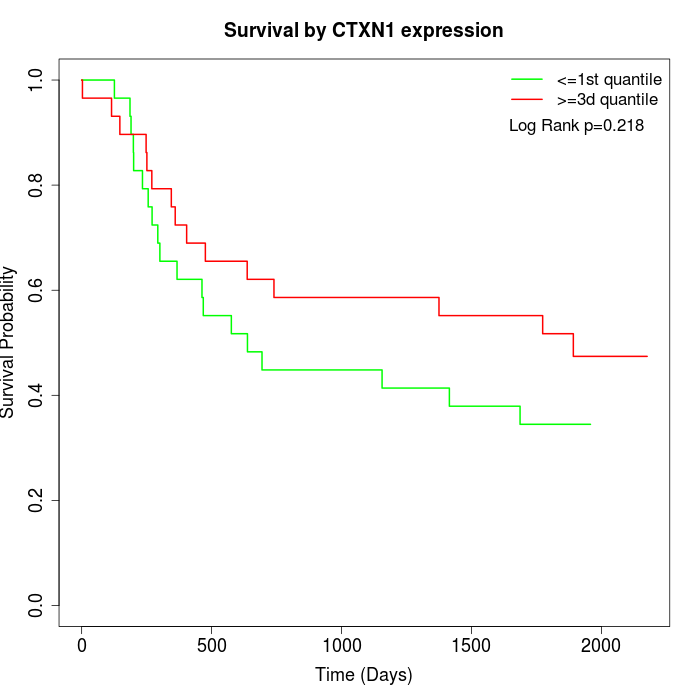
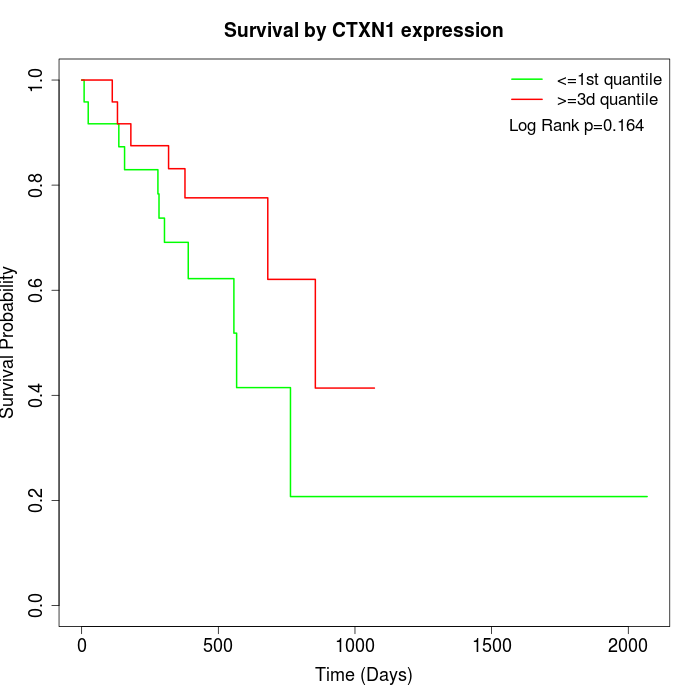
Survival by CTXN1 expression:
Note: Click image to view full size file.
Copy number change of CTXN1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTXN1 | 404217 | 4 | 3 | 23 | |
| GSE20123 | CTXN1 | 404217 | 3 | 2 | 25 | |
| GSE43470 | CTXN1 | 404217 | 2 | 7 | 34 | |
| GSE46452 | CTXN1 | 404217 | 47 | 1 | 11 | |
| GSE47630 | CTXN1 | 404217 | 5 | 7 | 28 | |
| GSE54993 | CTXN1 | 404217 | 16 | 3 | 51 | |
| GSE54994 | CTXN1 | 404217 | 6 | 14 | 33 | |
| GSE60625 | CTXN1 | 404217 | 9 | 0 | 2 | |
| GSE74703 | CTXN1 | 404217 | 2 | 4 | 30 | |
| GSE74704 | CTXN1 | 404217 | 0 | 2 | 18 | |
| TCGA | CTXN1 | 404217 | 9 | 20 | 67 |
Total number of gains: 103; Total number of losses: 63; Total Number of normals: 322.
Somatic mutations of CTXN1:
Generating mutation plots.
Highly correlated genes for CTXN1:
Showing top 20/183 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTXN1 | RASD2 | 0.762451 | 3 | 0 | 3 |
| CTXN1 | DMAP1 | 0.757385 | 3 | 0 | 3 |
| CTXN1 | ARHGAP4 | 0.750545 | 3 | 0 | 3 |
| CTXN1 | ZNF473 | 0.745407 | 3 | 0 | 3 |
| CTXN1 | LIG3 | 0.73279 | 3 | 0 | 3 |
| CTXN1 | BOD1 | 0.731995 | 3 | 0 | 3 |
| CTXN1 | PRSS36 | 0.721405 | 3 | 0 | 3 |
| CTXN1 | SPHK2 | 0.715414 | 3 | 0 | 3 |
| CTXN1 | SAE1 | 0.713128 | 3 | 0 | 3 |
| CTXN1 | TNPO2 | 0.711516 | 3 | 0 | 3 |
| CTXN1 | AMIGO1 | 0.710048 | 3 | 0 | 3 |
| CTXN1 | KCTD2 | 0.706115 | 3 | 0 | 3 |
| CTXN1 | SRF | 0.703951 | 3 | 0 | 3 |
| CTXN1 | SYNGR3 | 0.700836 | 4 | 0 | 4 |
| CTXN1 | ANKS3 | 0.698522 | 5 | 0 | 5 |
| CTXN1 | RBM17 | 0.697813 | 3 | 0 | 3 |
| CTXN1 | SPEG | 0.688599 | 3 | 0 | 3 |
| CTXN1 | ZNF74 | 0.687892 | 4 | 0 | 3 |
| CTXN1 | TNFRSF10C | 0.686772 | 3 | 0 | 3 |
| CTXN1 | MB | 0.682384 | 3 | 0 | 3 |
For details and further investigation, click here