| Full name: potassium channel tetramerization domain containing 4 | Alias Symbol: bA321C24.3 | ||
| Type: protein-coding gene | Cytoband: 13q14.12-q14.13 | ||
| Entrez ID: 386618 | HGNC ID: HGNC:23227 | Ensembl Gene: ENSG00000180332 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCTD4 | 386618 | 239787_at | -0.2205 | 0.2732 | |
| GSE26886 | KCTD4 | 386618 | 239787_at | -0.3600 | 0.0005 | |
| GSE45670 | KCTD4 | 386618 | 239787_at | -0.1990 | 0.0549 | |
| GSE53622 | KCTD4 | 386618 | 40042 | -0.8947 | 0.0000 | |
| GSE53624 | KCTD4 | 386618 | 40042 | -0.7049 | 0.0001 | |
| GSE63941 | KCTD4 | 386618 | 239787_at | 0.5257 | 0.6589 | |
| GSE77861 | KCTD4 | 386618 | 239787_at | -0.0760 | 0.3923 | |
| SRP133303 | KCTD4 | 386618 | RNAseq | 0.1485 | 0.4828 | |
| SRP193095 | KCTD4 | 386618 | RNAseq | -0.0726 | 0.6894 | |
| TCGA | KCTD4 | 386618 | RNAseq | -0.1444 | 0.7737 |
Upregulated datasets: 0; Downregulated datasets: 0.
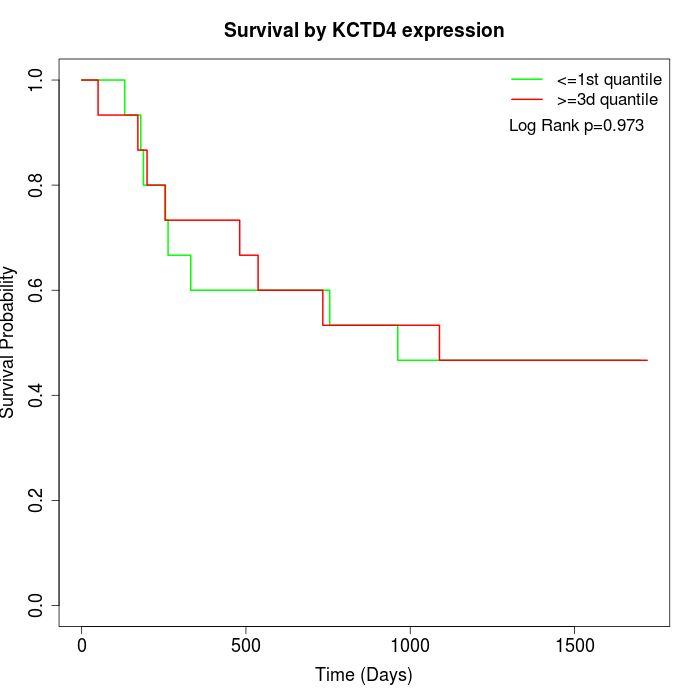
Survival by KCTD4 expression:
Note: Click image to view full size file.
Copy number change of KCTD4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD4 | 386618 | 0 | 13 | 17 | |
| GSE20123 | KCTD4 | 386618 | 0 | 12 | 18 | |
| GSE43470 | KCTD4 | 386618 | 3 | 13 | 27 | |
| GSE46452 | KCTD4 | 386618 | 0 | 33 | 26 | |
| GSE47630 | KCTD4 | 386618 | 2 | 27 | 11 | |
| GSE54993 | KCTD4 | 386618 | 12 | 2 | 56 | |
| GSE54994 | KCTD4 | 386618 | 1 | 16 | 36 | |
| GSE60625 | KCTD4 | 386618 | 0 | 3 | 8 | |
| GSE74703 | KCTD4 | 386618 | 3 | 10 | 23 | |
| GSE74704 | KCTD4 | 386618 | 0 | 9 | 11 | |
| TCGA | KCTD4 | 386618 | 8 | 39 | 49 |
Total number of gains: 29; Total number of losses: 177; Total Number of normals: 282.
Somatic mutations of KCTD4:
Generating mutation plots.
Highly correlated genes for KCTD4:
Showing top 20/347 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD4 | GBP6 | 0.724078 | 3 | 0 | 3 |
| KCTD4 | CLIC3 | 0.719356 | 3 | 0 | 3 |
| KCTD4 | MYO5B | 0.705905 | 3 | 0 | 3 |
| KCTD4 | CSTA | 0.705005 | 3 | 0 | 3 |
| KCTD4 | CEACAM5 | 0.69517 | 3 | 0 | 3 |
| KCTD4 | PHF7 | 0.694265 | 3 | 0 | 3 |
| KCTD4 | FUT3 | 0.689217 | 3 | 0 | 3 |
| KCTD4 | CYP2C18 | 0.688383 | 3 | 0 | 3 |
| KCTD4 | C10orf67 | 0.687618 | 3 | 0 | 3 |
| KCTD4 | PITX1 | 0.682939 | 3 | 0 | 3 |
| KCTD4 | TAAR5 | 0.679322 | 3 | 0 | 3 |
| KCTD4 | KRT33A | 0.677682 | 4 | 0 | 4 |
| KCTD4 | SNORA68 | 0.671647 | 3 | 0 | 3 |
| KCTD4 | SMIM5 | 0.669905 | 4 | 0 | 3 |
| KCTD4 | CES2 | 0.669455 | 3 | 0 | 3 |
| KCTD4 | RIOK3 | 0.661769 | 3 | 0 | 3 |
| KCTD4 | NDOR1 | 0.659623 | 3 | 0 | 3 |
| KCTD4 | TTC9 | 0.656057 | 4 | 0 | 3 |
| KCTD4 | ANKRD22 | 0.655964 | 3 | 0 | 3 |
| KCTD4 | SULT2B1 | 0.655037 | 3 | 0 | 3 |
For details and further investigation, click here