| Full name: kallikrein related peptidase 15 | Alias Symbol: HSRNASPH|ACO|prostinogen | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 55554 | HGNC ID: HGNC:20453 | Ensembl Gene: ENSG00000174562 | OMIM ID: 610601 |
| Drug and gene relationship at DGIdb | |||
Expression of KLK15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLK15 | 55554 | 234495_at | 0.0225 | 0.9454 | |
| GSE20347 | KLK15 | 55554 | 221462_x_at | -0.2198 | 0.0085 | |
| GSE23400 | KLK15 | 55554 | 221462_x_at | -0.1878 | 0.0000 | |
| GSE26886 | KLK15 | 55554 | 234495_at | 0.1565 | 0.3352 | |
| GSE29001 | KLK15 | 55554 | 221462_x_at | -0.1547 | 0.1428 | |
| GSE38129 | KLK15 | 55554 | 221462_x_at | -0.2035 | 0.0291 | |
| GSE45670 | KLK15 | 55554 | 234495_at | 0.0288 | 0.8444 | |
| GSE53622 | KLK15 | 55554 | 46446 | -0.0373 | 0.7313 | |
| GSE53624 | KLK15 | 55554 | 46446 | -0.3422 | 0.0013 | |
| GSE63941 | KLK15 | 55554 | 234495_at | 0.1944 | 0.3791 | |
| GSE77861 | KLK15 | 55554 | 234495_at | -0.0576 | 0.7328 | |
| TCGA | KLK15 | 55554 | RNAseq | -0.8771 | 0.4021 |
Upregulated datasets: 0; Downregulated datasets: 0.
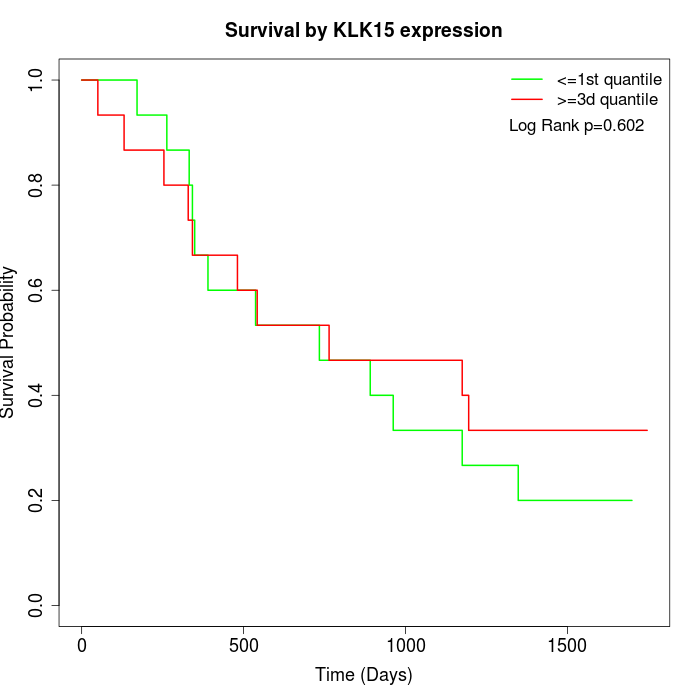
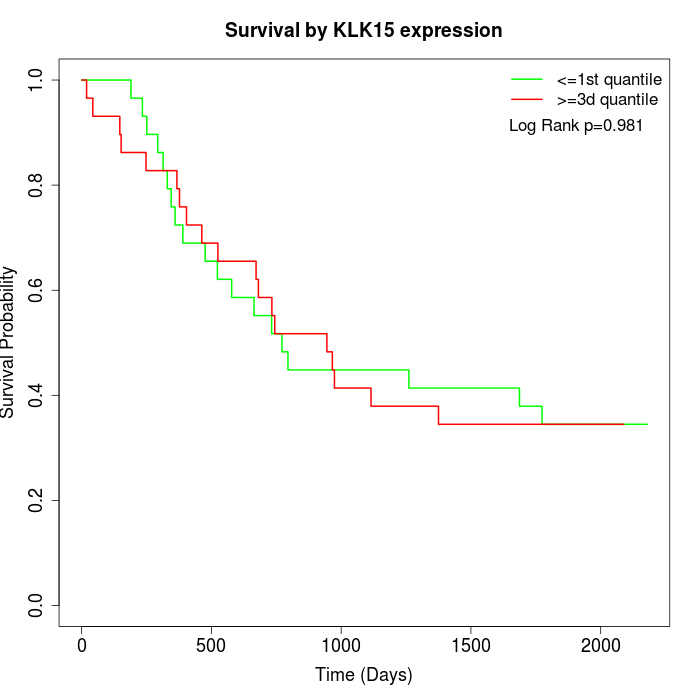
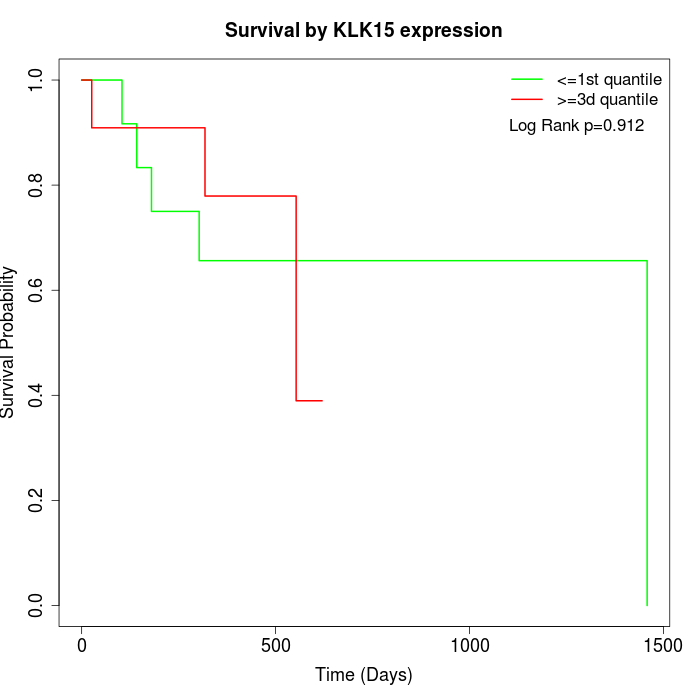
Survival by KLK15 expression:
Note: Click image to view full size file.
Copy number change of KLK15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLK15 | 55554 | 3 | 4 | 23 | |
| GSE20123 | KLK15 | 55554 | 3 | 3 | 24 | |
| GSE43470 | KLK15 | 55554 | 4 | 11 | 28 | |
| GSE46452 | KLK15 | 55554 | 45 | 1 | 13 | |
| GSE47630 | KLK15 | 55554 | 9 | 6 | 25 | |
| GSE54993 | KLK15 | 55554 | 17 | 4 | 49 | |
| GSE54994 | KLK15 | 55554 | 4 | 14 | 35 | |
| GSE60625 | KLK15 | 55554 | 9 | 0 | 2 | |
| GSE74703 | KLK15 | 55554 | 4 | 7 | 25 | |
| GSE74704 | KLK15 | 55554 | 3 | 1 | 16 | |
| TCGA | KLK15 | 55554 | 14 | 19 | 63 |
Total number of gains: 115; Total number of losses: 70; Total Number of normals: 303.
Somatic mutations of KLK15:
Generating mutation plots.
Highly correlated genes for KLK15:
Showing top 20/680 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLK15 | LINC00608 | 0.694789 | 4 | 0 | 4 |
| KLK15 | COL26A1 | 0.694671 | 3 | 0 | 3 |
| KLK15 | NOXA1 | 0.692467 | 3 | 0 | 3 |
| KLK15 | JSRP1 | 0.687076 | 4 | 0 | 4 |
| KLK15 | PAX5 | 0.682936 | 3 | 0 | 3 |
| KLK15 | SIK1 | 0.680339 | 4 | 0 | 3 |
| KLK15 | FAXDC2 | 0.676513 | 3 | 0 | 3 |
| KLK15 | ACRV1 | 0.668002 | 4 | 0 | 4 |
| KLK15 | GLYAT | 0.667827 | 4 | 0 | 4 |
| KLK15 | CCDC114 | 0.662079 | 3 | 0 | 3 |
| KLK15 | VWCE | 0.656278 | 3 | 0 | 3 |
| KLK15 | DLK1 | 0.655359 | 3 | 0 | 3 |
| KLK15 | LINC00475 | 0.652925 | 3 | 0 | 3 |
| KLK15 | LINC00482 | 0.647967 | 3 | 0 | 3 |
| KLK15 | MTAP | 0.646427 | 4 | 0 | 4 |
| KLK15 | HDAC10 | 0.646033 | 3 | 0 | 3 |
| KLK15 | PODXL2 | 0.641039 | 3 | 0 | 3 |
| KLK15 | VCX2 | 0.637391 | 5 | 0 | 5 |
| KLK15 | ITGB3 | 0.637292 | 3 | 0 | 3 |
| KLK15 | PNMA3 | 0.635432 | 6 | 0 | 6 |
For details and further investigation, click here