| Full name: lipocalin like 1 | Alias Symbol: FLJ45224 | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 401562 | HGNC ID: HGNC:34436 | Ensembl Gene: ENSG00000214402 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LCNL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCNL1 | 401562 | 1564431_a_at | -0.0563 | 0.8534 | |
| GSE26886 | LCNL1 | 401562 | 1560488_at | -0.1441 | 0.1657 | |
| GSE45670 | LCNL1 | 401562 | 1560488_at | 0.1693 | 0.0140 | |
| GSE53622 | LCNL1 | 401562 | 104725 | 0.4922 | 0.0000 | |
| GSE53624 | LCNL1 | 401562 | 104725 | 0.0789 | 0.2594 | |
| GSE63941 | LCNL1 | 401562 | 1564431_a_at | 0.2211 | 0.1287 | |
| GSE77861 | LCNL1 | 401562 | 1560488_at | -0.0283 | 0.7626 | |
| GSE97050 | LCNL1 | 401562 | A_33_P3390557 | 0.0936 | 0.6139 | |
| SRP219564 | LCNL1 | 401562 | RNAseq | -2.2645 | 0.0007 | |
| TCGA | LCNL1 | 401562 | RNAseq | -0.8121 | 0.2979 |
Upregulated datasets: 0; Downregulated datasets: 1.
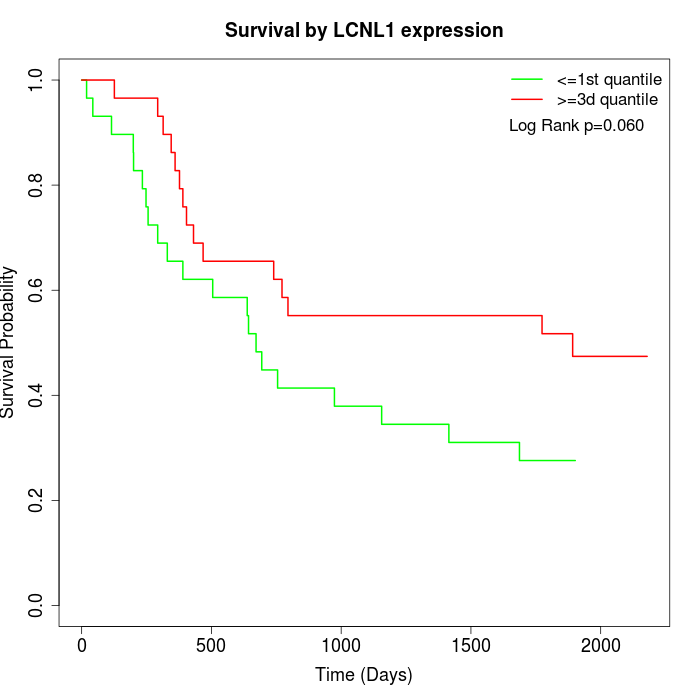
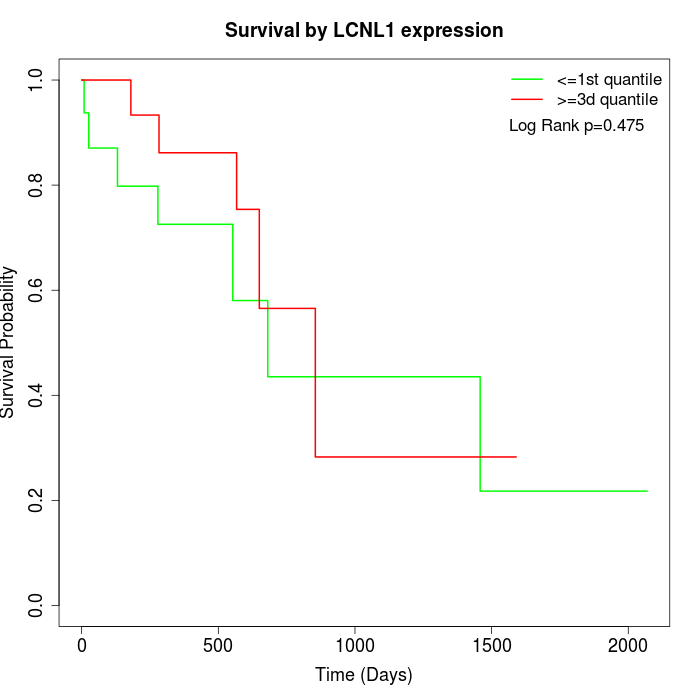
Survival by LCNL1 expression:
Note: Click image to view full size file.
Copy number change of LCNL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCNL1 | 401562 | 5 | 7 | 18 | |
| GSE20123 | LCNL1 | 401562 | 5 | 7 | 18 | |
| GSE43470 | LCNL1 | 401562 | 3 | 7 | 33 | |
| GSE46452 | LCNL1 | 401562 | 6 | 13 | 40 | |
| GSE47630 | LCNL1 | 401562 | 6 | 15 | 19 | |
| GSE54993 | LCNL1 | 401562 | 3 | 3 | 64 | |
| GSE54994 | LCNL1 | 401562 | 12 | 8 | 33 | |
| GSE60625 | LCNL1 | 401562 | 0 | 0 | 11 | |
| GSE74703 | LCNL1 | 401562 | 3 | 5 | 28 | |
| GSE74704 | LCNL1 | 401562 | 3 | 5 | 12 | |
| TCGA | LCNL1 | 401562 | 29 | 23 | 44 |
Total number of gains: 75; Total number of losses: 93; Total Number of normals: 320.
Somatic mutations of LCNL1:
Generating mutation plots.
Highly correlated genes for LCNL1:
Showing top 20/233 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCNL1 | OR52L1 | 0.733534 | 3 | 0 | 3 |
| LCNL1 | ROR1-AS1 | 0.69892 | 3 | 0 | 3 |
| LCNL1 | AK1 | 0.680588 | 4 | 0 | 4 |
| LCNL1 | FAM131C | 0.678968 | 3 | 0 | 3 |
| LCNL1 | SLC27A1 | 0.657294 | 3 | 0 | 3 |
| LCNL1 | RAX | 0.653963 | 3 | 0 | 3 |
| LCNL1 | KCTD19 | 0.653031 | 4 | 0 | 4 |
| LCNL1 | SPAG6 | 0.651742 | 3 | 0 | 3 |
| LCNL1 | TMPO-AS1 | 0.645512 | 3 | 0 | 3 |
| LCNL1 | TMEM174 | 0.642618 | 4 | 0 | 4 |
| LCNL1 | RPH3A | 0.641493 | 4 | 0 | 4 |
| LCNL1 | LRRC71 | 0.639097 | 4 | 0 | 4 |
| LCNL1 | BLACE | 0.633195 | 3 | 0 | 3 |
| LCNL1 | GRIK4 | 0.632672 | 3 | 0 | 3 |
| LCNL1 | GHSR | 0.630117 | 3 | 0 | 3 |
| LCNL1 | SPACA1 | 0.629794 | 4 | 0 | 3 |
| LCNL1 | BRPF1 | 0.626745 | 3 | 0 | 3 |
| LCNL1 | LRRC27 | 0.624969 | 5 | 0 | 5 |
| LCNL1 | TMEM82 | 0.622895 | 3 | 0 | 3 |
| LCNL1 | CYP46A1 | 0.622543 | 3 | 0 | 3 |
For details and further investigation, click here