| Full name: galectin 2 | Alias Symbol: HL14 | ||
| Type: protein-coding gene | Cytoband: 22q13.1 | ||
| Entrez ID: 3957 | HGNC ID: HGNC:6562 | Ensembl Gene: ENSG00000100079 | OMIM ID: 150571 |
| Drug and gene relationship at DGIdb | |||
Expression of LGALS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LGALS2 | 3957 | 208450_at | -0.3569 | 0.6662 | |
| GSE20347 | LGALS2 | 3957 | 208450_at | -0.2888 | 0.0289 | |
| GSE23400 | LGALS2 | 3957 | 208450_at | -0.1194 | 0.0691 | |
| GSE26886 | LGALS2 | 3957 | 208450_at | -0.4735 | 0.1595 | |
| GSE29001 | LGALS2 | 3957 | 208450_at | -0.3736 | 0.1240 | |
| GSE38129 | LGALS2 | 3957 | 208450_at | -0.1799 | 0.3375 | |
| GSE45670 | LGALS2 | 3957 | 208450_at | -0.3886 | 0.1002 | |
| GSE53622 | LGALS2 | 3957 | 12519 | -0.7929 | 0.0000 | |
| GSE53624 | LGALS2 | 3957 | 12519 | -1.2051 | 0.0000 | |
| GSE63941 | LGALS2 | 3957 | 208450_at | 0.3263 | 0.0120 | |
| GSE77861 | LGALS2 | 3957 | 208450_at | -0.0976 | 0.5113 | |
| GSE97050 | LGALS2 | 3957 | A_23_P120902 | 0.2064 | 0.6146 | |
| SRP064894 | LGALS2 | 3957 | RNAseq | 0.0789 | 0.8598 | |
| SRP133303 | LGALS2 | 3957 | RNAseq | -1.0341 | 0.0001 | |
| SRP159526 | LGALS2 | 3957 | RNAseq | -1.3196 | 0.1710 | |
| SRP193095 | LGALS2 | 3957 | RNAseq | -0.5410 | 0.0538 | |
| SRP219564 | LGALS2 | 3957 | RNAseq | -0.2569 | 0.6735 | |
| TCGA | LGALS2 | 3957 | RNAseq | -0.7579 | 0.1093 |
Upregulated datasets: 0; Downregulated datasets: 2.
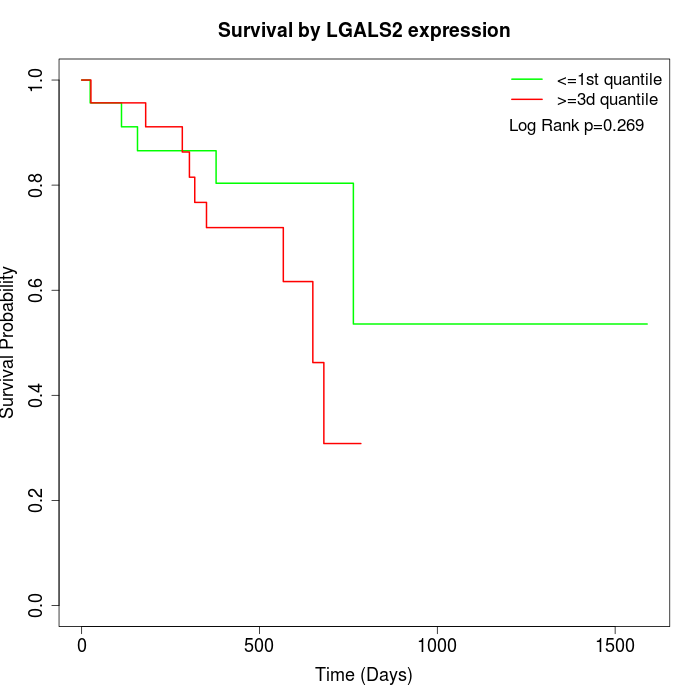
Survival by LGALS2 expression:
Note: Click image to view full size file.
Copy number change of LGALS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LGALS2 | 3957 | 6 | 5 | 19 | |
| GSE20123 | LGALS2 | 3957 | 6 | 4 | 20 | |
| GSE43470 | LGALS2 | 3957 | 4 | 6 | 33 | |
| GSE46452 | LGALS2 | 3957 | 32 | 2 | 25 | |
| GSE47630 | LGALS2 | 3957 | 9 | 4 | 27 | |
| GSE54993 | LGALS2 | 3957 | 3 | 6 | 61 | |
| GSE54994 | LGALS2 | 3957 | 9 | 8 | 36 | |
| GSE60625 | LGALS2 | 3957 | 5 | 0 | 6 | |
| GSE74703 | LGALS2 | 3957 | 4 | 4 | 28 | |
| GSE74704 | LGALS2 | 3957 | 3 | 2 | 15 | |
| TCGA | LGALS2 | 3957 | 27 | 15 | 54 |
Total number of gains: 108; Total number of losses: 56; Total Number of normals: 324.
Somatic mutations of LGALS2:
Generating mutation plots.
Highly correlated genes for LGALS2:
Showing top 20/302 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LGALS2 | MPEG1 | 0.767666 | 6 | 0 | 6 |
| LGALS2 | C16orf54 | 0.740651 | 6 | 0 | 6 |
| LGALS2 | GIMAP5 | 0.740151 | 4 | 0 | 3 |
| LGALS2 | P2RY8 | 0.739785 | 4 | 0 | 4 |
| LGALS2 | GIMAP7 | 0.736257 | 7 | 0 | 6 |
| LGALS2 | SNX20 | 0.726518 | 6 | 0 | 6 |
| LGALS2 | ARHGAP15 | 0.722277 | 3 | 0 | 3 |
| LGALS2 | C1orf162 | 0.718161 | 6 | 0 | 6 |
| LGALS2 | TAGAP | 0.714494 | 5 | 0 | 5 |
| LGALS2 | HLA-DOA | 0.708375 | 5 | 0 | 5 |
| LGALS2 | SPOCK2 | 0.707722 | 7 | 0 | 7 |
| LGALS2 | XPNPEP2 | 0.704481 | 4 | 0 | 4 |
| LGALS2 | LILRA4 | 0.703164 | 3 | 0 | 3 |
| LGALS2 | ARHGAP9 | 0.686321 | 6 | 0 | 5 |
| LGALS2 | RCSD1 | 0.684449 | 6 | 0 | 6 |
| LGALS2 | GIMAP6 | 0.68112 | 9 | 0 | 8 |
| LGALS2 | CNTFR | 0.679948 | 3 | 0 | 3 |
| LGALS2 | GNGT2 | 0.679617 | 6 | 0 | 6 |
| LGALS2 | CD2 | 0.677578 | 10 | 0 | 8 |
| LGALS2 | COL4A4 | 0.674745 | 4 | 0 | 3 |
For details and further investigation, click here