| Full name: leukocyte immunoglobulin like receptor A4 | Alias Symbol: ILT7|CD85g | ||
| Type: protein-coding gene | Cytoband: 19q13.42 | ||
| Entrez ID: 23547 | HGNC ID: HGNC:15503 | Ensembl Gene: ENSG00000239961 | OMIM ID: 607517 |
| Drug and gene relationship at DGIdb | |||
LILRA4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation |
Expression of LILRA4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LILRA4 | 23547 | 210313_at | -0.0323 | 0.8980 | |
| GSE20347 | LILRA4 | 23547 | 210313_at | 0.2050 | 0.0190 | |
| GSE23400 | LILRA4 | 23547 | 210313_at | -0.0557 | 0.1388 | |
| GSE26886 | LILRA4 | 23547 | 210313_at | 0.0149 | 0.9546 | |
| GSE29001 | LILRA4 | 23547 | 210313_at | 0.0825 | 0.7275 | |
| GSE38129 | LILRA4 | 23547 | 210313_at | -0.0882 | 0.3870 | |
| GSE45670 | LILRA4 | 23547 | 210313_at | 0.1367 | 0.1728 | |
| GSE63941 | LILRA4 | 23547 | 210313_at | 0.0700 | 0.7136 | |
| GSE77861 | LILRA4 | 23547 | 210313_at | -0.1334 | 0.2493 | |
| GSE97050 | LILRA4 | 23547 | A_23_P90497 | 1.0517 | 0.1113 | |
| TCGA | LILRA4 | 23547 | RNAseq | 0.4674 | 0.5391 |
Upregulated datasets: 0; Downregulated datasets: 0.
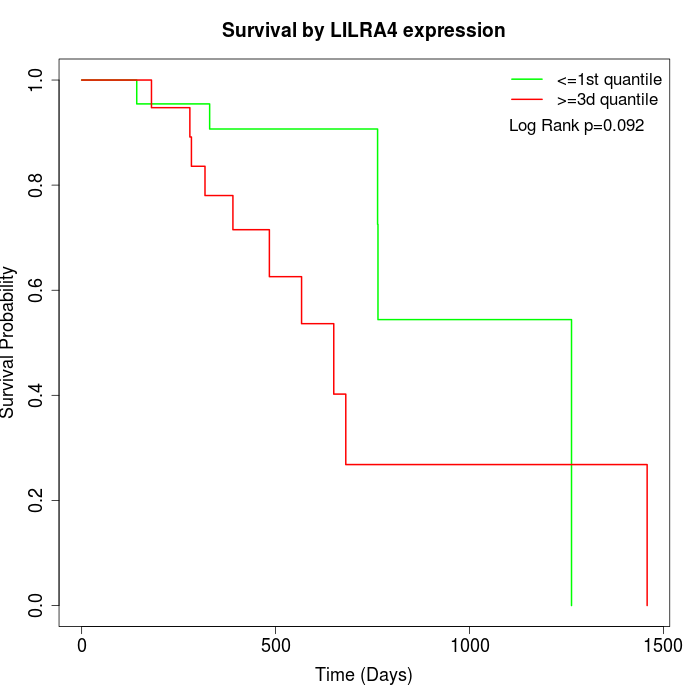
Survival by LILRA4 expression:
Note: Click image to view full size file.
Copy number change of LILRA4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LILRA4 | 23547 | 3 | 4 | 23 | |
| GSE20123 | LILRA4 | 23547 | 3 | 3 | 24 | |
| GSE43470 | LILRA4 | 23547 | 2 | 11 | 30 | |
| GSE46452 | LILRA4 | 23547 | 45 | 1 | 13 | |
| GSE47630 | LILRA4 | 23547 | 8 | 6 | 26 | |
| GSE54993 | LILRA4 | 23547 | 17 | 4 | 49 | |
| GSE54994 | LILRA4 | 23547 | 4 | 13 | 36 | |
| GSE60625 | LILRA4 | 23547 | 9 | 0 | 2 | |
| GSE74703 | LILRA4 | 23547 | 2 | 7 | 27 | |
| GSE74704 | LILRA4 | 23547 | 3 | 1 | 16 | |
| TCGA | LILRA4 | 23547 | 19 | 15 | 62 |
Total number of gains: 115; Total number of losses: 65; Total Number of normals: 308.
Somatic mutations of LILRA4:
Generating mutation plots.
Highly correlated genes for LILRA4:
Showing top 20/688 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LILRA4 | SAMD3 | 0.763226 | 3 | 0 | 3 |
| LILRA4 | TNFSF8 | 0.750453 | 5 | 0 | 5 |
| LILRA4 | NR5A2 | 0.723063 | 3 | 0 | 3 |
| LILRA4 | MAST2 | 0.721487 | 3 | 0 | 3 |
| LILRA4 | BTBD2 | 0.719275 | 3 | 0 | 3 |
| LILRA4 | SARDH | 0.710924 | 4 | 0 | 4 |
| LILRA4 | GATA6 | 0.706359 | 3 | 0 | 3 |
| LILRA4 | SAMD9L | 0.703538 | 3 | 0 | 3 |
| LILRA4 | LGALS2 | 0.703164 | 3 | 0 | 3 |
| LILRA4 | GTF2H3 | 0.697958 | 3 | 0 | 3 |
| LILRA4 | ATG10 | 0.697242 | 5 | 0 | 5 |
| LILRA4 | CR1L | 0.692276 | 3 | 0 | 3 |
| LILRA4 | ZNF358 | 0.689033 | 3 | 0 | 3 |
| LILRA4 | KIR2DL5A | 0.687089 | 5 | 0 | 5 |
| LILRA4 | VEGFA | 0.686512 | 3 | 0 | 3 |
| LILRA4 | LIPF | 0.68192 | 3 | 0 | 3 |
| LILRA4 | SMARCD1 | 0.678787 | 3 | 0 | 3 |
| LILRA4 | CLDN2 | 0.677779 | 3 | 0 | 3 |
| LILRA4 | SCML4 | 0.673113 | 3 | 0 | 3 |
| LILRA4 | KHDRBS2 | 0.672739 | 6 | 0 | 6 |
For details and further investigation, click here