| Full name: LIM homeobox 1 | Alias Symbol: LIM-1|LIM1 | ||
| Type: protein-coding gene | Cytoband: 17q12 | ||
| Entrez ID: 3975 | HGNC ID: HGNC:6593 | Ensembl Gene: ENSG00000273706 | OMIM ID: 601999 |
| Drug and gene relationship at DGIdb | |||
Expression of LHX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LHX1 | 3975 | 206230_at | 0.0876 | 0.6256 | |
| GSE20347 | LHX1 | 3975 | 206230_at | 0.0663 | 0.3206 | |
| GSE23400 | LHX1 | 3975 | 206230_at | 0.0007 | 0.9792 | |
| GSE26886 | LHX1 | 3975 | 206230_at | 0.4944 | 0.0022 | |
| GSE29001 | LHX1 | 3975 | 206230_at | 0.1199 | 0.3547 | |
| GSE38129 | LHX1 | 3975 | 206230_at | 0.0184 | 0.8279 | |
| GSE45670 | LHX1 | 3975 | 206230_at | 0.1772 | 0.2289 | |
| GSE53622 | LHX1 | 3975 | 3425 | 0.5377 | 0.0014 | |
| GSE53624 | LHX1 | 3975 | 3425 | 0.6572 | 0.0012 | |
| GSE63941 | LHX1 | 3975 | 206230_at | 0.3220 | 0.0998 | |
| GSE77861 | LHX1 | 3975 | 206230_at | -0.0526 | 0.6878 | |
| GSE97050 | LHX1 | 3975 | A_33_P3409854 | 0.2732 | 0.5189 | |
| TCGA | LHX1 | 3975 | RNAseq | 1.9473 | 0.0453 |
Upregulated datasets: 1; Downregulated datasets: 0.
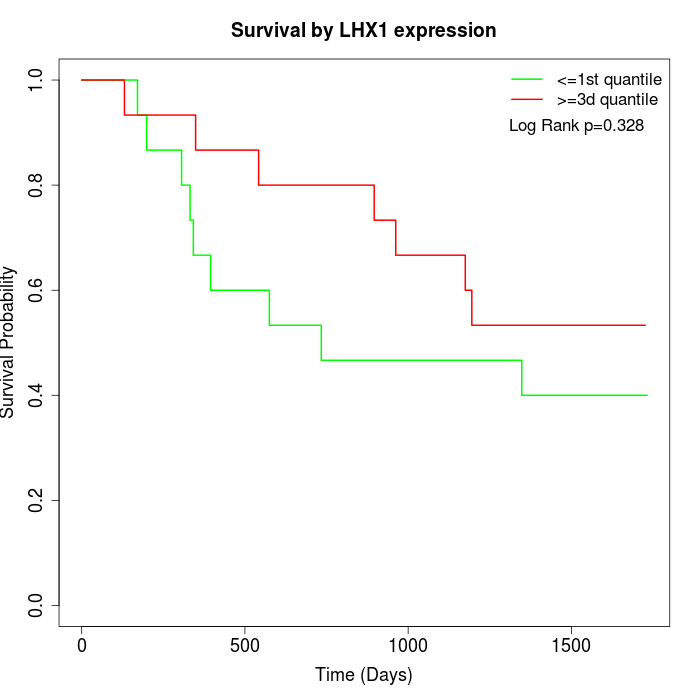
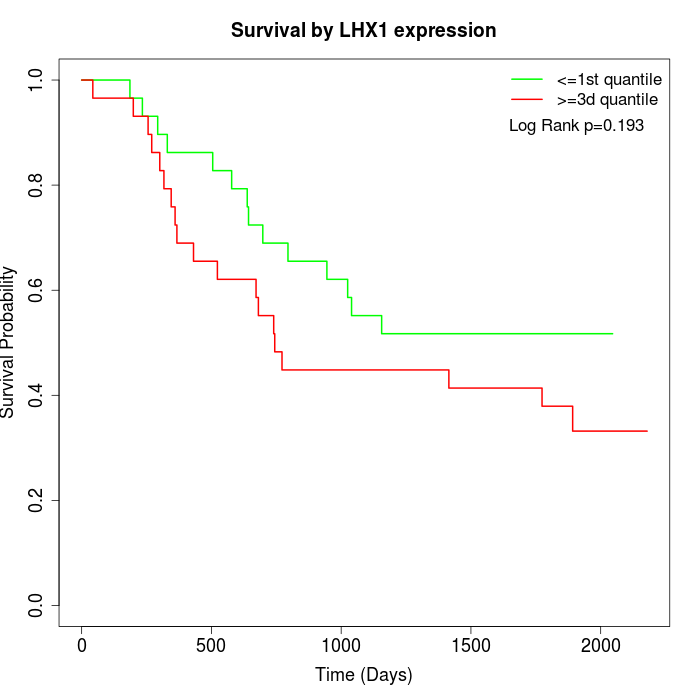
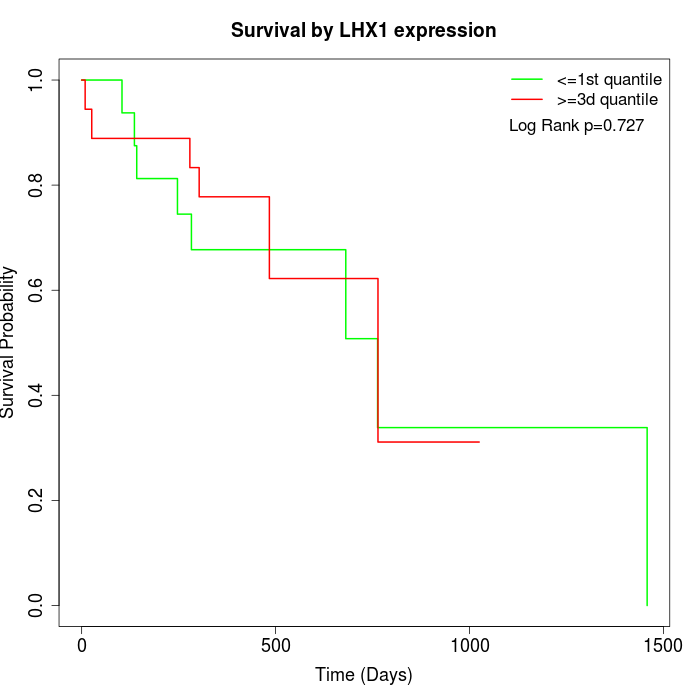
Survival by LHX1 expression:
Note: Click image to view full size file.
Copy number change of LHX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LHX1 | 3975 | 5 | 1 | 24 | |
| GSE20123 | LHX1 | 3975 | 5 | 1 | 24 | |
| GSE43470 | LHX1 | 3975 | 0 | 2 | 41 | |
| GSE46452 | LHX1 | 3975 | 35 | 0 | 24 | |
| GSE47630 | LHX1 | 3975 | 8 | 1 | 31 | |
| GSE54993 | LHX1 | 3975 | 3 | 3 | 64 | |
| GSE54994 | LHX1 | 3975 | 8 | 6 | 39 | |
| GSE60625 | LHX1 | 3975 | 4 | 0 | 7 | |
| GSE74703 | LHX1 | 3975 | 0 | 1 | 35 | |
| GSE74704 | LHX1 | 3975 | 3 | 1 | 16 | |
| TCGA | LHX1 | 3975 | 19 | 9 | 68 |
Total number of gains: 90; Total number of losses: 25; Total Number of normals: 373.
Somatic mutations of LHX1:
Generating mutation plots.
Highly correlated genes for LHX1:
Showing top 20/372 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LHX1 | RLN3 | 0.7529 | 3 | 0 | 3 |
| LHX1 | NPW | 0.729274 | 3 | 0 | 3 |
| LHX1 | SPATA2L | 0.721103 | 4 | 0 | 4 |
| LHX1 | TMEM102 | 0.718119 | 4 | 0 | 4 |
| LHX1 | PPIH | 0.711674 | 3 | 0 | 3 |
| LHX1 | STAC3 | 0.702037 | 3 | 0 | 3 |
| LHX1 | PCSK9 | 0.700933 | 3 | 0 | 3 |
| LHX1 | MTFP1 | 0.699657 | 5 | 0 | 5 |
| LHX1 | TEX13B | 0.698207 | 3 | 0 | 3 |
| LHX1 | DNAJB8 | 0.69751 | 3 | 0 | 3 |
| LHX1 | POLD1 | 0.694998 | 3 | 0 | 3 |
| LHX1 | PERM1 | 0.689174 | 4 | 0 | 4 |
| LHX1 | RASL10B | 0.687315 | 4 | 0 | 3 |
| LHX1 | GPR150 | 0.686068 | 4 | 0 | 4 |
| LHX1 | DOHH | 0.680834 | 4 | 0 | 4 |
| LHX1 | CACNA1A | 0.680579 | 4 | 0 | 3 |
| LHX1 | PIK3CD | 0.678147 | 3 | 0 | 3 |
| LHX1 | SCUBE1 | 0.674474 | 4 | 0 | 4 |
| LHX1 | JSRP1 | 0.671229 | 4 | 0 | 3 |
| LHX1 | ZWINT | 0.661976 | 3 | 0 | 3 |
For details and further investigation, click here