| Full name: LIM domain kinase 1 | Alias Symbol: LIMK | ||
| Type: protein-coding gene | Cytoband: 7q11.23 | ||
| Entrez ID: 3984 | HGNC ID: HGNC:6613 | Ensembl Gene: ENSG00000106683 | OMIM ID: 601329 |
| Drug and gene relationship at DGIdb | |||
LIMK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04666 | Fc gamma R-mediated phagocytosis | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of LIMK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIMK1 | 3984 | 204357_s_at | 1.5332 | 0.0182 | |
| GSE20347 | LIMK1 | 3984 | 204357_s_at | 0.3764 | 0.0001 | |
| GSE23400 | LIMK1 | 3984 | 204357_s_at | 0.3904 | 0.0000 | |
| GSE26886 | LIMK1 | 3984 | 204357_s_at | 0.3811 | 0.0198 | |
| GSE29001 | LIMK1 | 3984 | 204357_s_at | 0.6203 | 0.0002 | |
| GSE38129 | LIMK1 | 3984 | 204357_s_at | 0.6530 | 0.0000 | |
| GSE45670 | LIMK1 | 3984 | 204357_s_at | 0.3833 | 0.0075 | |
| GSE53622 | LIMK1 | 3984 | 34642 | 1.2365 | 0.0000 | |
| GSE53624 | LIMK1 | 3984 | 34642 | 1.5873 | 0.0000 | |
| GSE63941 | LIMK1 | 3984 | 204357_s_at | -0.6126 | 0.1574 | |
| GSE77861 | LIMK1 | 3984 | 204357_s_at | 0.3102 | 0.0841 | |
| GSE97050 | LIMK1 | 3984 | A_23_P215461 | 1.1002 | 0.0871 | |
| SRP007169 | LIMK1 | 3984 | RNAseq | 1.7371 | 0.0031 | |
| SRP008496 | LIMK1 | 3984 | RNAseq | 1.7634 | 0.0000 | |
| SRP064894 | LIMK1 | 3984 | RNAseq | 1.5180 | 0.0000 | |
| SRP133303 | LIMK1 | 3984 | RNAseq | 1.4870 | 0.0000 | |
| SRP159526 | LIMK1 | 3984 | RNAseq | 1.2790 | 0.0000 | |
| SRP193095 | LIMK1 | 3984 | RNAseq | 1.5429 | 0.0000 | |
| SRP219564 | LIMK1 | 3984 | RNAseq | 1.7900 | 0.0007 | |
| TCGA | LIMK1 | 3984 | RNAseq | 0.4155 | 0.0000 |
Upregulated datasets: 10; Downregulated datasets: 0.
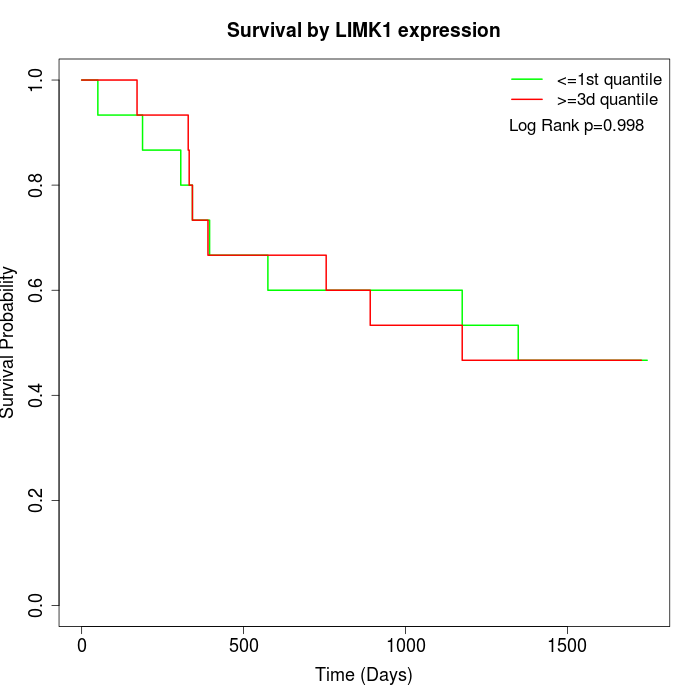
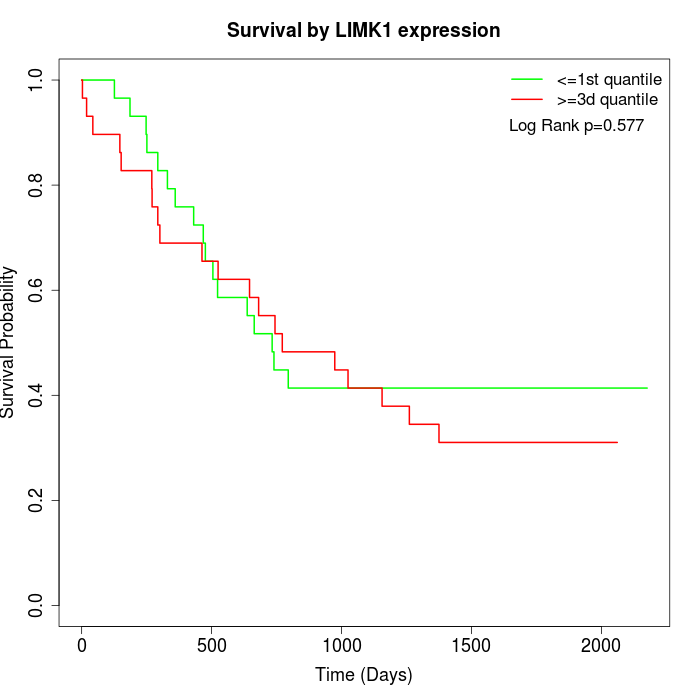
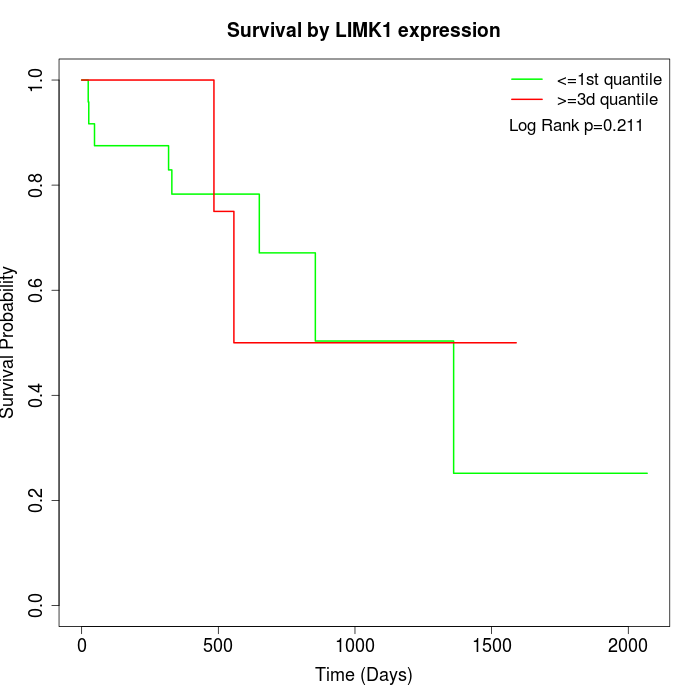
Survival by LIMK1 expression:
Note: Click image to view full size file.
Copy number change of LIMK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIMK1 | 3984 | 14 | 1 | 15 | |
| GSE20123 | LIMK1 | 3984 | 14 | 1 | 15 | |
| GSE43470 | LIMK1 | 3984 | 3 | 2 | 38 | |
| GSE46452 | LIMK1 | 3984 | 12 | 0 | 47 | |
| GSE47630 | LIMK1 | 3984 | 7 | 2 | 31 | |
| GSE54993 | LIMK1 | 3984 | 2 | 7 | 61 | |
| GSE54994 | LIMK1 | 3984 | 13 | 3 | 37 | |
| GSE60625 | LIMK1 | 3984 | 0 | 0 | 11 | |
| GSE74703 | LIMK1 | 3984 | 3 | 1 | 32 | |
| GSE74704 | LIMK1 | 3984 | 8 | 1 | 11 | |
| TCGA | LIMK1 | 3984 | 45 | 7 | 44 |
Total number of gains: 121; Total number of losses: 25; Total Number of normals: 342.
Somatic mutations of LIMK1:
Generating mutation plots.
Highly correlated genes for LIMK1:
Showing top 20/1923 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIMK1 | PYCR2 | 0.803903 | 3 | 0 | 3 |
| LIMK1 | RNF4 | 0.785494 | 3 | 0 | 3 |
| LIMK1 | GORAB | 0.757309 | 5 | 0 | 5 |
| LIMK1 | EBNA1BP2 | 0.757069 | 3 | 0 | 3 |
| LIMK1 | DOCK6 | 0.754162 | 4 | 0 | 4 |
| LIMK1 | RUNX2 | 0.751632 | 3 | 0 | 3 |
| LIMK1 | PRDM15 | 0.749705 | 3 | 0 | 3 |
| LIMK1 | FAM89A | 0.748715 | 3 | 0 | 3 |
| LIMK1 | FMNL3 | 0.748664 | 5 | 0 | 5 |
| LIMK1 | PHRF1 | 0.745812 | 4 | 0 | 4 |
| LIMK1 | SPON2 | 0.744024 | 3 | 0 | 3 |
| LIMK1 | ZNF707 | 0.74368 | 3 | 0 | 3 |
| LIMK1 | CENPBD1 | 0.742993 | 3 | 0 | 3 |
| LIMK1 | PROX1 | 0.739247 | 3 | 0 | 3 |
| LIMK1 | KRTCAP2 | 0.736023 | 4 | 0 | 4 |
| LIMK1 | PDIA5 | 0.735506 | 3 | 0 | 3 |
| LIMK1 | KIAA0930 | 0.732965 | 11 | 0 | 10 |
| LIMK1 | BOLA3 | 0.726553 | 6 | 0 | 6 |
| LIMK1 | RNF213 | 0.724551 | 3 | 0 | 3 |
| LIMK1 | SLC19A1 | 0.723694 | 3 | 0 | 3 |
For details and further investigation, click here