| Full name: ankyrin repeat and SOCS box containing 5 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q34.2 | ||
| Entrez ID: 140458 | HGNC ID: HGNC:17180 | Ensembl Gene: ENSG00000164122 | OMIM ID: 615050 |
| Drug and gene relationship at DGIdb | |||
Expression of ASB5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ASB5 | 140458 | 235503_at | -1.8509 | 0.1497 | |
| GSE26886 | ASB5 | 140458 | 235503_at | 0.1047 | 0.2090 | |
| GSE45670 | ASB5 | 140458 | 235503_at | -2.5472 | 0.0000 | |
| GSE53622 | ASB5 | 140458 | 11944 | -2.8720 | 0.0000 | |
| GSE53624 | ASB5 | 140458 | 11944 | -2.9564 | 0.0000 | |
| GSE63941 | ASB5 | 140458 | 235503_at | -0.5885 | 0.0003 | |
| GSE77861 | ASB5 | 140458 | 235503_at | -0.0337 | 0.7271 | |
| GSE97050 | ASB5 | 140458 | A_23_P144326 | -1.2736 | 0.1399 | |
| SRP064894 | ASB5 | 140458 | RNAseq | -0.6027 | 0.3397 | |
| SRP133303 | ASB5 | 140458 | RNAseq | -3.3311 | 0.0000 | |
| SRP159526 | ASB5 | 140458 | RNAseq | -1.2564 | 0.2568 | |
| TCGA | ASB5 | 140458 | RNAseq | -1.3666 | 0.1067 |
Upregulated datasets: 0; Downregulated datasets: 4.
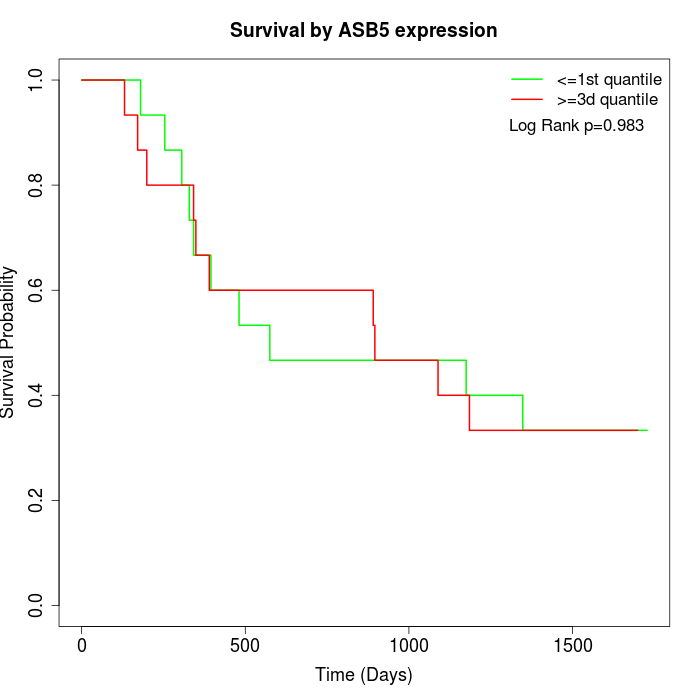
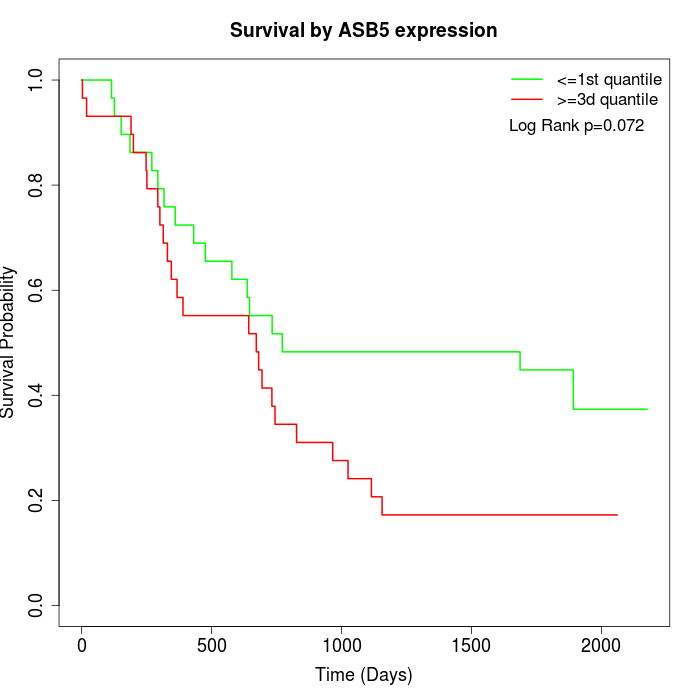
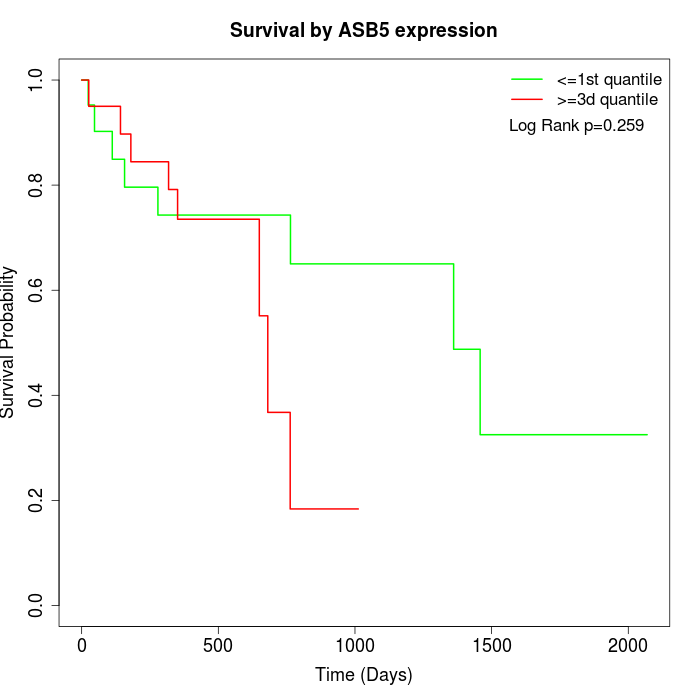
Survival by ASB5 expression:
Note: Click image to view full size file.
Copy number change of ASB5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ASB5 | 140458 | 1 | 10 | 19 | |
| GSE20123 | ASB5 | 140458 | 1 | 10 | 19 | |
| GSE43470 | ASB5 | 140458 | 0 | 14 | 29 | |
| GSE46452 | ASB5 | 140458 | 1 | 36 | 22 | |
| GSE47630 | ASB5 | 140458 | 0 | 22 | 18 | |
| GSE54993 | ASB5 | 140458 | 10 | 0 | 60 | |
| GSE54994 | ASB5 | 140458 | 2 | 12 | 39 | |
| GSE60625 | ASB5 | 140458 | 0 | 0 | 11 | |
| GSE74703 | ASB5 | 140458 | 0 | 12 | 24 | |
| GSE74704 | ASB5 | 140458 | 0 | 5 | 15 | |
| TCGA | ASB5 | 140458 | 10 | 36 | 50 |
Total number of gains: 25; Total number of losses: 157; Total Number of normals: 306.
Somatic mutations of ASB5:
Generating mutation plots.
Highly correlated genes for ASB5:
Showing top 20/1030 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ASB5 | SORCS1 | 0.868742 | 4 | 0 | 4 |
| ASB5 | CASQ2 | 0.865254 | 6 | 0 | 6 |
| ASB5 | MYOCD | 0.865161 | 6 | 0 | 6 |
| ASB5 | SLC2A4 | 0.863983 | 3 | 0 | 3 |
| ASB5 | BHMT2 | 0.855531 | 6 | 0 | 6 |
| ASB5 | PCP4 | 0.847665 | 5 | 0 | 5 |
| ASB5 | ATP1A2 | 0.846662 | 5 | 0 | 5 |
| ASB5 | PGM5 | 0.841679 | 5 | 0 | 5 |
| ASB5 | AFF3 | 0.839925 | 3 | 0 | 3 |
| ASB5 | SLITRK3 | 0.836936 | 5 | 0 | 5 |
| ASB5 | FENDRR | 0.834268 | 5 | 0 | 5 |
| ASB5 | NEGR1 | 0.830294 | 6 | 0 | 6 |
| ASB5 | CKMT2 | 0.829661 | 5 | 0 | 5 |
| ASB5 | SYNC | 0.826682 | 4 | 0 | 4 |
| ASB5 | BCHE | 0.821504 | 5 | 0 | 5 |
| ASB5 | C1QTNF7 | 0.820632 | 5 | 0 | 5 |
| ASB5 | ITGA1 | 0.819933 | 5 | 0 | 5 |
| ASB5 | HOXA4 | 0.817118 | 3 | 0 | 3 |
| ASB5 | DES | 0.817016 | 5 | 0 | 5 |
| ASB5 | KCNB1 | 0.815554 | 5 | 0 | 5 |
For details and further investigation, click here