| Full name: methyl-CpG binding protein 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xq28 | ||
| Entrez ID: 4204 | HGNC ID: HGNC:6990 | Ensembl Gene: ENSG00000169057 | OMIM ID: 300005 |
| Drug and gene relationship at DGIdb | |||
Expression of MECP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MECP2 | 4204 | 202616_s_at | -0.1824 | 0.7427 | |
| GSE20347 | MECP2 | 4204 | 202616_s_at | 0.1224 | 0.3622 | |
| GSE23400 | MECP2 | 4204 | 202616_s_at | -0.1120 | 0.1139 | |
| GSE26886 | MECP2 | 4204 | 202618_s_at | 0.4215 | 0.0004 | |
| GSE29001 | MECP2 | 4204 | 202616_s_at | -0.0080 | 0.9782 | |
| GSE38129 | MECP2 | 4204 | 202616_s_at | -0.1711 | 0.3482 | |
| GSE45670 | MECP2 | 4204 | 202618_s_at | -0.0304 | 0.8150 | |
| GSE53622 | MECP2 | 4204 | 23577 | 0.1615 | 0.0022 | |
| GSE53624 | MECP2 | 4204 | 23577 | 0.1430 | 0.0007 | |
| GSE63941 | MECP2 | 4204 | 202616_s_at | -0.1158 | 0.7429 | |
| GSE77861 | MECP2 | 4204 | 202617_s_at | 0.2342 | 0.2012 | |
| GSE97050 | MECP2 | 4204 | A_33_P3317211 | -0.4793 | 0.1657 | |
| SRP007169 | MECP2 | 4204 | RNAseq | -0.1779 | 0.7386 | |
| SRP008496 | MECP2 | 4204 | RNAseq | 0.1585 | 0.6363 | |
| SRP064894 | MECP2 | 4204 | RNAseq | 0.5292 | 0.0235 | |
| SRP133303 | MECP2 | 4204 | RNAseq | 0.2221 | 0.2729 | |
| SRP159526 | MECP2 | 4204 | RNAseq | 0.2451 | 0.2336 | |
| SRP193095 | MECP2 | 4204 | RNAseq | 0.2665 | 0.0138 | |
| SRP219564 | MECP2 | 4204 | RNAseq | 0.1468 | 0.6402 | |
| TCGA | MECP2 | 4204 | RNAseq | -0.1651 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
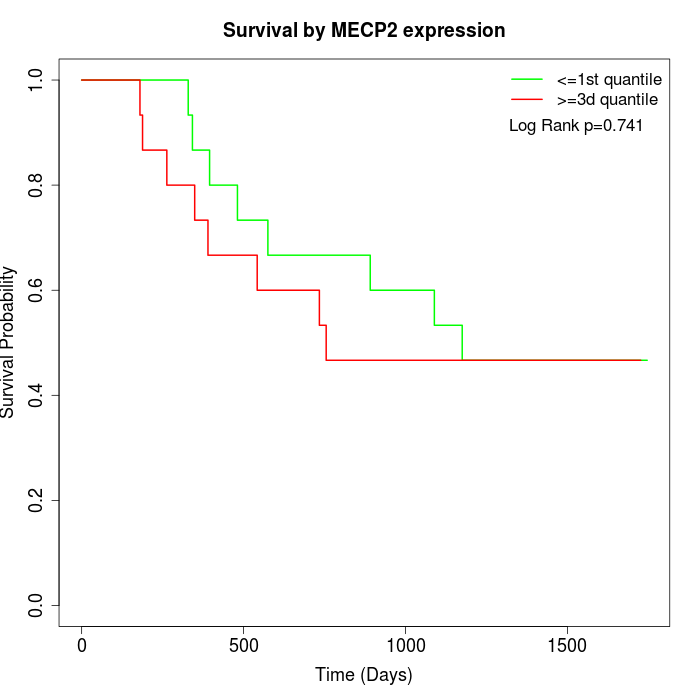
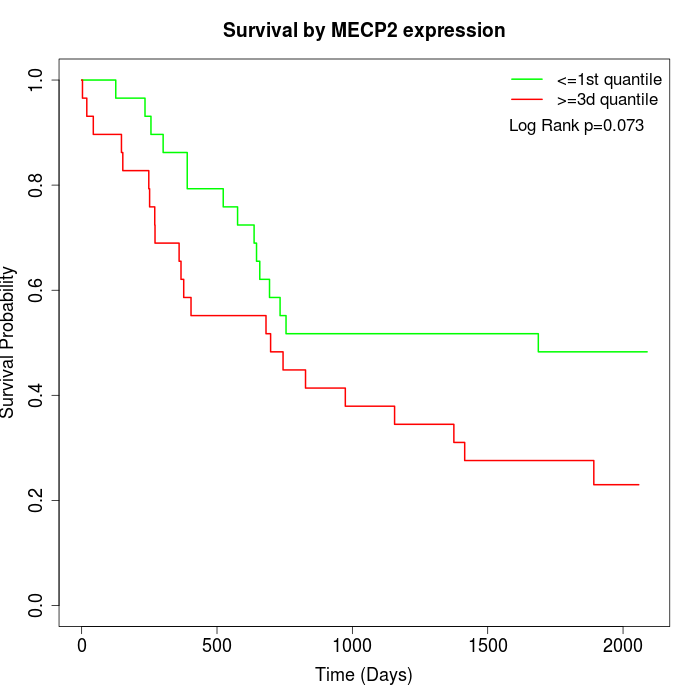
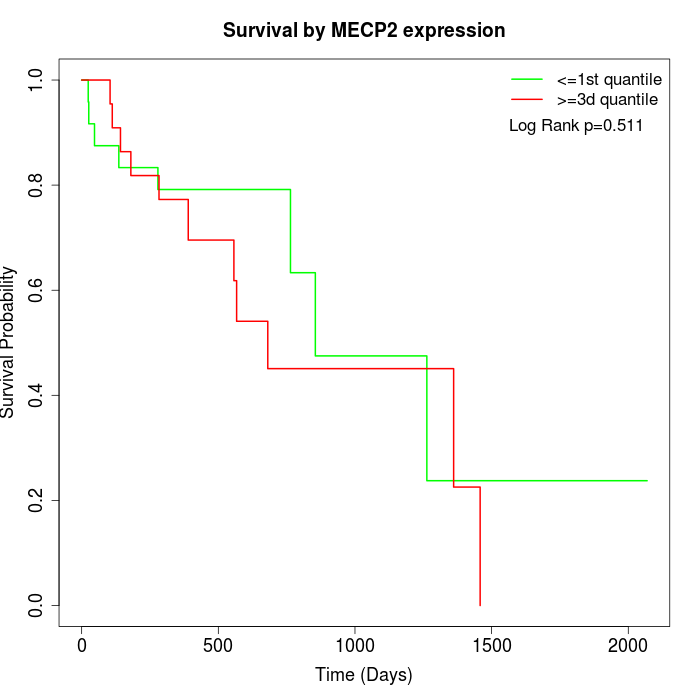
Survival by MECP2 expression:
Note: Click image to view full size file.
Copy number change of MECP2:
No record found for this gene.
Somatic mutations of MECP2:
Generating mutation plots.
Highly correlated genes for MECP2:
Showing top 20/769 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MECP2 | HYI | 0.819177 | 3 | 0 | 3 |
| MECP2 | ITGA7 | 0.792675 | 4 | 0 | 4 |
| MECP2 | LIMS2 | 0.779642 | 5 | 0 | 5 |
| MECP2 | PYGM | 0.774809 | 4 | 0 | 4 |
| MECP2 | KCNMB1 | 0.772894 | 4 | 0 | 4 |
| MECP2 | DDX42 | 0.77282 | 3 | 0 | 3 |
| MECP2 | BHMT2 | 0.772575 | 4 | 0 | 4 |
| MECP2 | UBAC1 | 0.762443 | 3 | 0 | 3 |
| MECP2 | MEOX2 | 0.760171 | 5 | 0 | 5 |
| MECP2 | NCAM1 | 0.759528 | 4 | 0 | 4 |
| MECP2 | FRAS1 | 0.758981 | 3 | 0 | 3 |
| MECP2 | INMT | 0.758308 | 3 | 0 | 3 |
| MECP2 | TCEAL1 | 0.755759 | 4 | 0 | 4 |
| MECP2 | AQP1 | 0.751652 | 4 | 0 | 4 |
| MECP2 | PI16 | 0.748035 | 3 | 0 | 3 |
| MECP2 | PPP1R12B | 0.746589 | 4 | 0 | 4 |
| MECP2 | PLCL1 | 0.745945 | 4 | 0 | 4 |
| MECP2 | SYNC | 0.740944 | 4 | 0 | 4 |
| MECP2 | PPARGC1A | 0.737283 | 4 | 0 | 4 |
| MECP2 | RAB9B | 0.734199 | 3 | 0 | 3 |
For details and further investigation, click here