| Full name: potassium calcium-activated channel subfamily M alpha 1 | Alias Symbol: KCa1.1|mSLO1 | ||
| Type: protein-coding gene | Cytoband: 10q22.3 | ||
| Entrez ID: 3778 | HGNC ID: HGNC:6284 | Ensembl Gene: ENSG00000156113 | OMIM ID: 600150 |
| Drug and gene relationship at DGIdb | |||
KCNMA1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04270 | Vascular smooth muscle contraction | |
| hsa04924 | Renin secretion | |
| hsa04970 | Salivary secretion | |
| hsa04972 | Pancreatic secretion |
Expression of KCNMA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNMA1 | 3778 | 221584_s_at | -2.1538 | 0.1683 | |
| GSE20347 | KCNMA1 | 3778 | 221584_s_at | -0.9250 | 0.0189 | |
| GSE23400 | KCNMA1 | 3778 | 221584_s_at | -0.7751 | 0.0004 | |
| GSE26886 | KCNMA1 | 3778 | 221584_s_at | 1.8820 | 0.0001 | |
| GSE29001 | KCNMA1 | 3778 | 221584_s_at | -0.7768 | 0.2832 | |
| GSE38129 | KCNMA1 | 3778 | 221584_s_at | -1.3584 | 0.0134 | |
| GSE45670 | KCNMA1 | 3778 | 221584_s_at | -2.8582 | 0.0000 | |
| GSE53622 | KCNMA1 | 3778 | 61584 | -1.9913 | 0.0000 | |
| GSE53624 | KCNMA1 | 3778 | 28569 | -1.7992 | 0.0000 | |
| GSE63941 | KCNMA1 | 3778 | 221584_s_at | -3.6930 | 0.0055 | |
| GSE77861 | KCNMA1 | 3778 | 221584_s_at | 0.0204 | 0.9572 | |
| GSE97050 | KCNMA1 | 3778 | A_23_P60727 | -1.9264 | 0.1204 | |
| SRP007169 | KCNMA1 | 3778 | RNAseq | 2.5783 | 0.0011 | |
| SRP008496 | KCNMA1 | 3778 | RNAseq | 3.4095 | 0.0000 | |
| SRP064894 | KCNMA1 | 3778 | RNAseq | 0.2515 | 0.6019 | |
| SRP133303 | KCNMA1 | 3778 | RNAseq | 0.2109 | 0.7198 | |
| SRP159526 | KCNMA1 | 3778 | RNAseq | -0.9080 | 0.1057 | |
| SRP193095 | KCNMA1 | 3778 | RNAseq | 0.6670 | 0.0161 | |
| SRP219564 | KCNMA1 | 3778 | RNAseq | 0.0394 | 0.9657 | |
| TCGA | KCNMA1 | 3778 | RNAseq | -0.7078 | 0.0014 |
Upregulated datasets: 3; Downregulated datasets: 5.
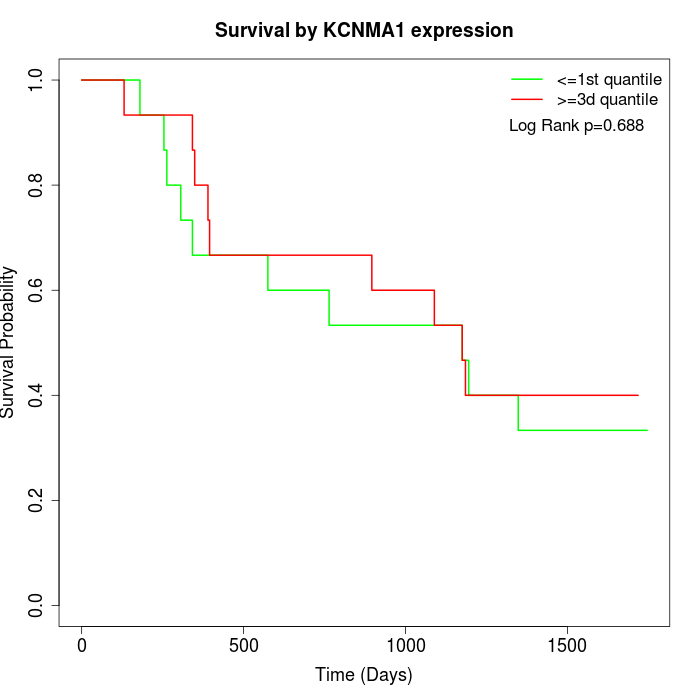
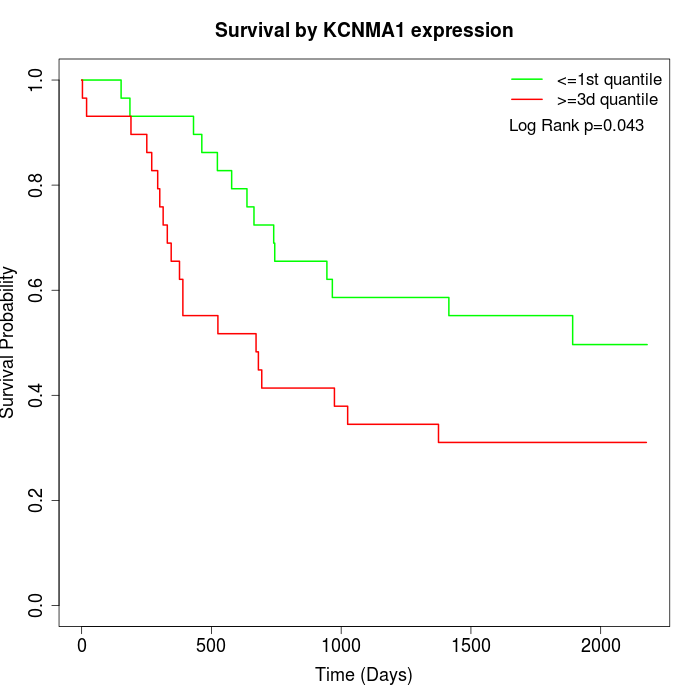
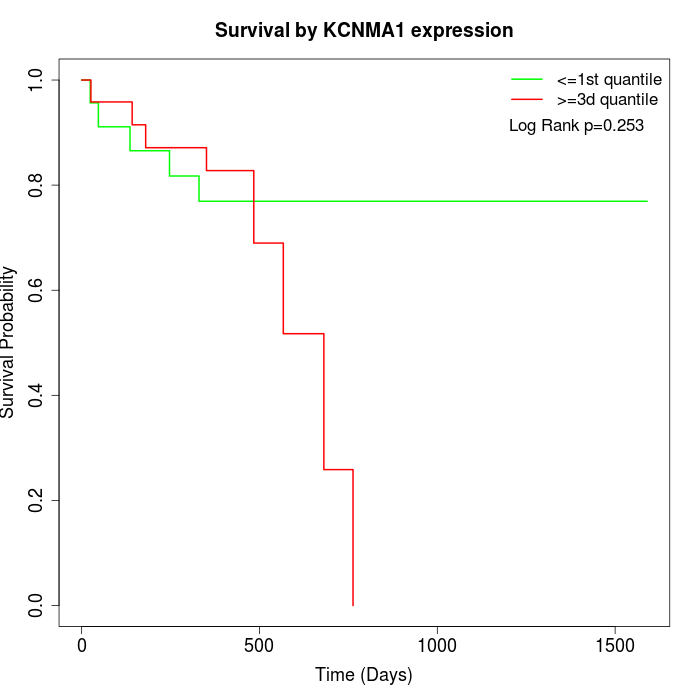
Survival by KCNMA1 expression:
Note: Click image to view full size file.
Copy number change of KCNMA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNMA1 | 3778 | 2 | 5 | 23 | |
| GSE20123 | KCNMA1 | 3778 | 2 | 4 | 24 | |
| GSE43470 | KCNMA1 | 3778 | 1 | 7 | 35 | |
| GSE46452 | KCNMA1 | 3778 | 0 | 11 | 48 | |
| GSE47630 | KCNMA1 | 3778 | 2 | 14 | 24 | |
| GSE54993 | KCNMA1 | 3778 | 7 | 0 | 63 | |
| GSE54994 | KCNMA1 | 3778 | 1 | 12 | 40 | |
| GSE60625 | KCNMA1 | 3778 | 0 | 0 | 11 | |
| GSE74703 | KCNMA1 | 3778 | 1 | 4 | 31 | |
| GSE74704 | KCNMA1 | 3778 | 1 | 2 | 17 | |
| TCGA | KCNMA1 | 3778 | 10 | 24 | 62 |
Total number of gains: 27; Total number of losses: 83; Total Number of normals: 378.
Somatic mutations of KCNMA1:
Generating mutation plots.
Highly correlated genes for KCNMA1:
Showing top 20/1143 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNMA1 | UBXN10 | 0.821672 | 3 | 0 | 3 |
| KCNMA1 | KANK2 | 0.812207 | 9 | 0 | 8 |
| KCNMA1 | PLN | 0.805901 | 12 | 0 | 12 |
| KCNMA1 | BHMT2 | 0.804705 | 9 | 0 | 9 |
| KCNMA1 | LMOD1 | 0.804314 | 12 | 0 | 12 |
| KCNMA1 | SORCS1 | 0.801135 | 4 | 0 | 4 |
| KCNMA1 | ANGPTL1 | 0.796489 | 6 | 0 | 6 |
| KCNMA1 | NEGR1 | 0.794347 | 6 | 0 | 6 |
| KCNMA1 | AOC3 | 0.793821 | 11 | 0 | 10 |
| KCNMA1 | CHRDL1 | 0.79152 | 9 | 0 | 8 |
| KCNMA1 | ASB5 | 0.783757 | 6 | 0 | 6 |
| KCNMA1 | ACTG2 | 0.783547 | 12 | 0 | 12 |
| KCNMA1 | LIMS2 | 0.782896 | 10 | 0 | 9 |
| KCNMA1 | ZNF347 | 0.780509 | 3 | 0 | 3 |
| KCNMA1 | PRUNE2 | 0.778855 | 12 | 0 | 12 |
| KCNMA1 | MSRB3 | 0.772672 | 7 | 0 | 6 |
| KCNMA1 | NCAM1 | 0.772577 | 8 | 0 | 8 |
| KCNMA1 | FLNC | 0.769559 | 12 | 0 | 11 |
| KCNMA1 | LONRF2 | 0.768875 | 5 | 0 | 5 |
| KCNMA1 | PGR | 0.768186 | 5 | 0 | 5 |
For details and further investigation, click here