| Full name: long intergenic non-protein coding RNA 865 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 10q23.31 | ||
| Entrez ID: 643529 | HGNC ID: HGNC:45170 | Ensembl Gene: ENSG00000232229 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00865:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00865 | 643529 | 232239_at | 0.0358 | 0.9703 | |
| GSE26886 | LINC00865 | 643529 | 232239_at | 0.3019 | 0.4200 | |
| GSE45670 | LINC00865 | 643529 | 232239_at | -0.1059 | 0.7438 | |
| GSE53622 | LINC00865 | 643529 | 46817 | 0.0088 | 0.9639 | |
| GSE53624 | LINC00865 | 643529 | 46817 | 0.0960 | 0.4238 | |
| GSE63941 | LINC00865 | 643529 | 232239_at | -1.9312 | 0.0001 | |
| GSE77861 | LINC00865 | 643529 | 232239_at | 0.0732 | 0.7303 | |
| SRP064894 | LINC00865 | 643529 | RNAseq | -0.2914 | 0.3777 | |
| SRP133303 | LINC00865 | 643529 | RNAseq | 0.1668 | 0.5244 | |
| SRP159526 | LINC00865 | 643529 | RNAseq | 0.8165 | 0.0827 | |
| SRP219564 | LINC00865 | 643529 | RNAseq | -0.4821 | 0.4502 |
Upregulated datasets: 0; Downregulated datasets: 1.
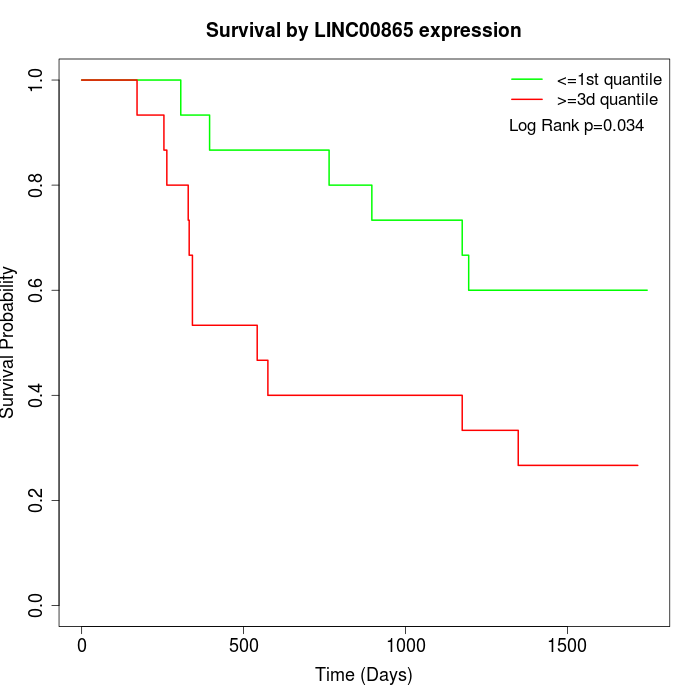
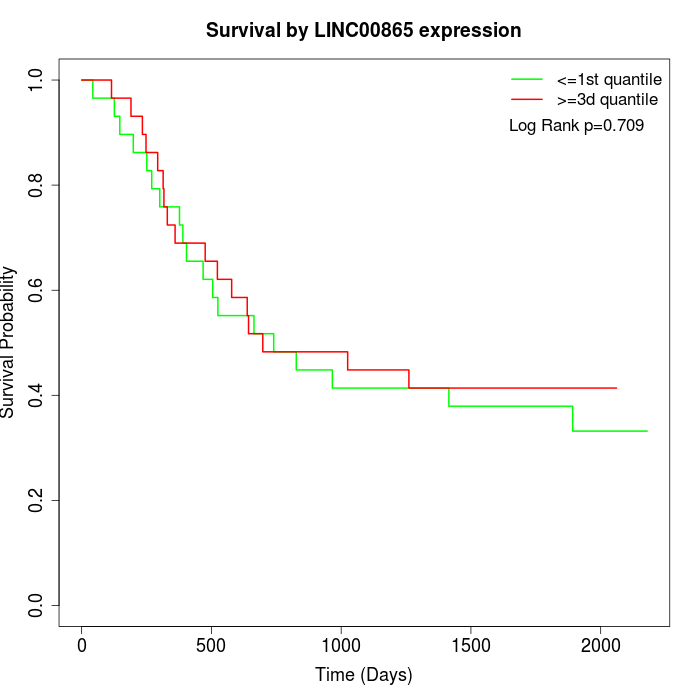
Survival by LINC00865 expression:
Note: Click image to view full size file.
Copy number change of LINC00865:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00865 | 643529 | 2 | 7 | 21 | |
| GSE20123 | LINC00865 | 643529 | 2 | 6 | 22 | |
| GSE43470 | LINC00865 | 643529 | 0 | 8 | 35 | |
| GSE46452 | LINC00865 | 643529 | 0 | 11 | 48 | |
| GSE47630 | LINC00865 | 643529 | 2 | 14 | 24 | |
| GSE54993 | LINC00865 | 643529 | 7 | 0 | 63 | |
| GSE54994 | LINC00865 | 643529 | 1 | 11 | 41 | |
| GSE60625 | LINC00865 | 643529 | 0 | 0 | 11 | |
| GSE74703 | LINC00865 | 643529 | 0 | 6 | 30 | |
| GSE74704 | LINC00865 | 643529 | 1 | 4 | 15 | |
| TCGA | LINC00865 | 643529 | 7 | 27 | 62 |
Total number of gains: 22; Total number of losses: 94; Total Number of normals: 372.
Somatic mutations of LINC00865:
Generating mutation plots.
Highly correlated genes for LINC00865:
Showing top 20/62 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00865 | CLIC6 | 0.6941 | 4 | 0 | 4 |
| LINC00865 | ACTG2 | 0.686311 | 3 | 0 | 3 |
| LINC00865 | TRDN | 0.671101 | 3 | 0 | 3 |
| LINC00865 | ZNF443 | 0.656973 | 3 | 0 | 3 |
| LINC00865 | ZNF610 | 0.631076 | 3 | 0 | 3 |
| LINC00865 | CLEC1A | 0.62083 | 3 | 0 | 3 |
| LINC00865 | MAGED1 | 0.620196 | 4 | 0 | 3 |
| LINC00865 | SPON1 | 0.616839 | 3 | 0 | 3 |
| LINC00865 | ERP27 | 0.615185 | 3 | 0 | 3 |
| LINC00865 | TCTN1 | 0.61493 | 3 | 0 | 3 |
| LINC00865 | HERC2 | 0.613769 | 3 | 0 | 3 |
| LINC00865 | SYNPO2 | 0.61201 | 4 | 0 | 3 |
| LINC00865 | CHST14 | 0.611996 | 4 | 0 | 3 |
| LINC00865 | GSPT2 | 0.602301 | 5 | 0 | 4 |
| LINC00865 | ZNF322 | 0.601672 | 4 | 0 | 3 |
| LINC00865 | SCAF8 | 0.600712 | 3 | 0 | 3 |
| LINC00865 | USP51 | 0.598577 | 4 | 0 | 3 |
| LINC00865 | PDE4B | 0.596479 | 3 | 0 | 3 |
| LINC00865 | NR3C2 | 0.595188 | 4 | 0 | 4 |
| LINC00865 | BAALC | 0.594117 | 3 | 0 | 3 |
For details and further investigation, click here