| Full name: serine protease 35 | Alias Symbol: MGC46520|dJ223E3.1 | ||
| Type: protein-coding gene | Cytoband: 6q14.2 | ||
| Entrez ID: 167681 | HGNC ID: HGNC:21387 | Ensembl Gene: ENSG00000146250 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of PRSS35:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRSS35 | 167681 | 235874_at | 0.0156 | 0.9491 | |
| GSE26886 | PRSS35 | 167681 | 235874_at | 0.1413 | 0.2098 | |
| GSE45670 | PRSS35 | 167681 | 235874_at | -0.3089 | 0.0586 | |
| GSE53622 | PRSS35 | 167681 | 22154 | -1.0501 | 0.0000 | |
| GSE53624 | PRSS35 | 167681 | 22154 | -0.5772 | 0.0027 | |
| GSE63941 | PRSS35 | 167681 | 235874_at | -3.9885 | 0.0000 | |
| GSE77861 | PRSS35 | 167681 | 235874_at | -0.0527 | 0.6952 | |
| SRP133303 | PRSS35 | 167681 | RNAseq | -0.2141 | 0.5035 | |
| SRP159526 | PRSS35 | 167681 | RNAseq | 0.0983 | 0.8557 | |
| TCGA | PRSS35 | 167681 | RNAseq | -1.0249 | 0.0281 |
Upregulated datasets: 0; Downregulated datasets: 3.
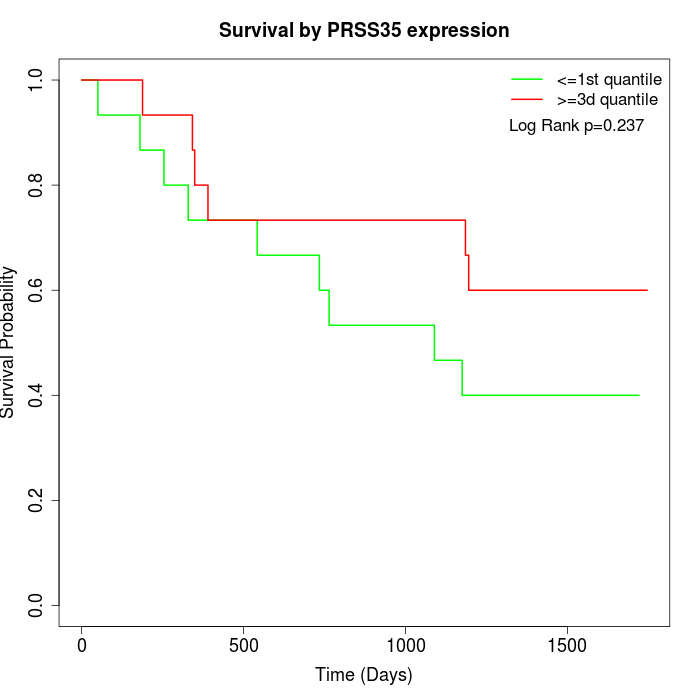
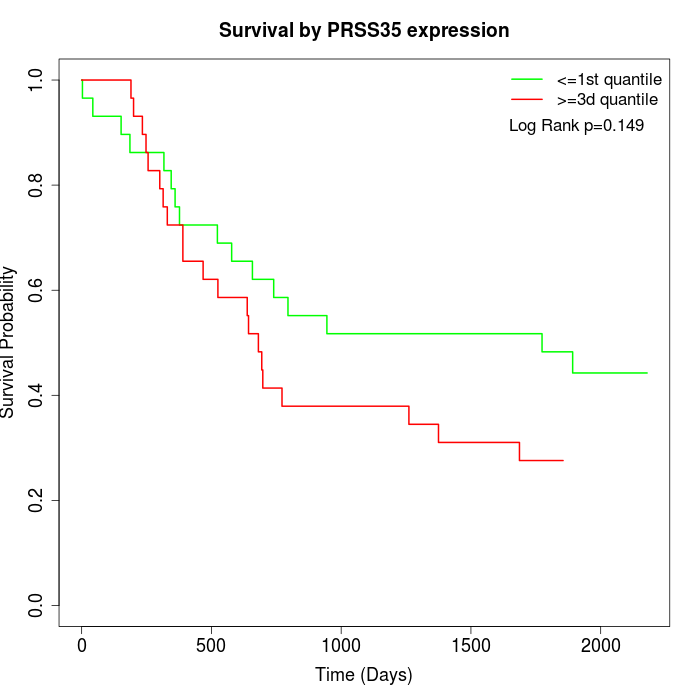
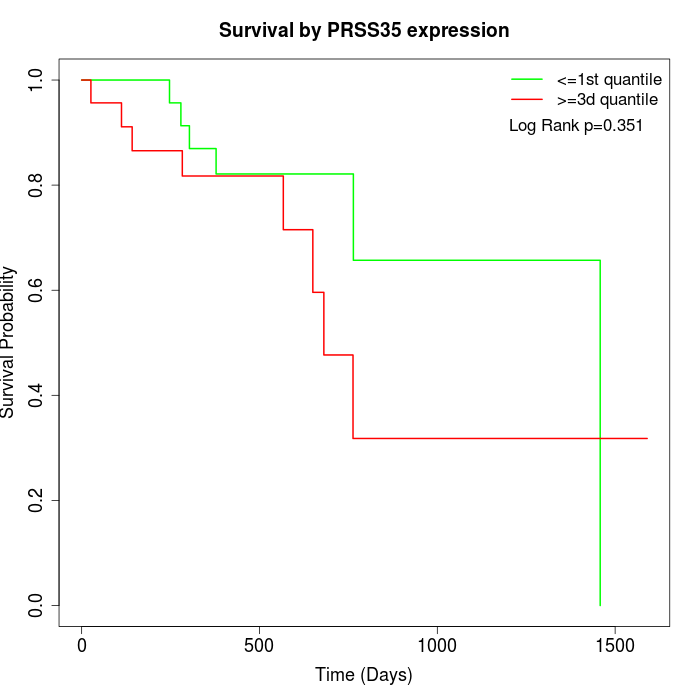
Survival by PRSS35 expression:
Note: Click image to view full size file.
Copy number change of PRSS35:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRSS35 | 167681 | 1 | 4 | 25 | |
| GSE20123 | PRSS35 | 167681 | 1 | 3 | 26 | |
| GSE43470 | PRSS35 | 167681 | 2 | 1 | 40 | |
| GSE46452 | PRSS35 | 167681 | 2 | 11 | 46 | |
| GSE47630 | PRSS35 | 167681 | 8 | 6 | 26 | |
| GSE54993 | PRSS35 | 167681 | 3 | 2 | 65 | |
| GSE54994 | PRSS35 | 167681 | 8 | 8 | 37 | |
| GSE60625 | PRSS35 | 167681 | 0 | 1 | 10 | |
| GSE74703 | PRSS35 | 167681 | 2 | 1 | 33 | |
| GSE74704 | PRSS35 | 167681 | 0 | 2 | 18 | |
| TCGA | PRSS35 | 167681 | 6 | 21 | 69 |
Total number of gains: 33; Total number of losses: 60; Total Number of normals: 395.
Somatic mutations of PRSS35:
Generating mutation plots.
Highly correlated genes for PRSS35:
Showing top 20/158 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRSS35 | WFDC1 | 0.703655 | 3 | 0 | 3 |
| PRSS35 | LINC00968 | 0.700699 | 4 | 0 | 4 |
| PRSS35 | HSD11B1 | 0.699771 | 3 | 0 | 3 |
| PRSS35 | SCN9A | 0.692564 | 3 | 0 | 3 |
| PRSS35 | FBN2 | 0.690628 | 3 | 0 | 3 |
| PRSS35 | STK32B | 0.689861 | 5 | 0 | 4 |
| PRSS35 | TARSL2 | 0.682523 | 3 | 0 | 3 |
| PRSS35 | SLC16A4 | 0.67716 | 4 | 0 | 4 |
| PRSS35 | ADAMTS5 | 0.673677 | 4 | 0 | 4 |
| PRSS35 | GALNT15 | 0.671462 | 3 | 0 | 3 |
| PRSS35 | NLGN1 | 0.669889 | 4 | 0 | 3 |
| PRSS35 | TMEM100 | 0.66922 | 4 | 0 | 4 |
| PRSS35 | IL33 | 0.667493 | 3 | 0 | 3 |
| PRSS35 | VAT1L | 0.662616 | 4 | 0 | 4 |
| PRSS35 | PKD2 | 0.657679 | 3 | 0 | 3 |
| PRSS35 | GCSAML | 0.655749 | 3 | 0 | 3 |
| PRSS35 | ARHGAP20 | 0.655523 | 3 | 0 | 3 |
| PRSS35 | MAP3K3 | 0.65358 | 3 | 0 | 3 |
| PRSS35 | GNG11 | 0.653425 | 4 | 0 | 4 |
| PRSS35 | AOX1 | 0.652321 | 3 | 0 | 3 |
For details and further investigation, click here