| Full name: long intergenic non-protein coding RNA 1191 | Alias Symbol: VIN|lnc-ACTR3 | ||
| Type: non-coding RNA | Cytoband: 2q14.1 | ||
| Entrez ID: 440900 | HGNC ID: HGNC:49595 | Ensembl Gene: ENSG00000234199 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC01191:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC01191 | 440900 | 230796_at | 0.0081 | 0.9864 | |
| GSE26886 | LINC01191 | 440900 | 230796_at | 0.4121 | 0.0137 | |
| GSE45670 | LINC01191 | 440900 | 230796_at | 0.0074 | 0.9615 | |
| GSE53622 | LINC01191 | 440900 | 105895 | 0.5424 | 0.0056 | |
| GSE53624 | LINC01191 | 440900 | 105895 | 0.4045 | 0.0055 | |
| GSE63941 | LINC01191 | 440900 | 230796_at | -0.0967 | 0.4996 | |
| GSE77861 | LINC01191 | 440900 | 230796_at | -0.0171 | 0.9137 | |
| SRP133303 | LINC01191 | 440900 | RNAseq | -0.3853 | 0.0579 |
Upregulated datasets: 0; Downregulated datasets: 0.
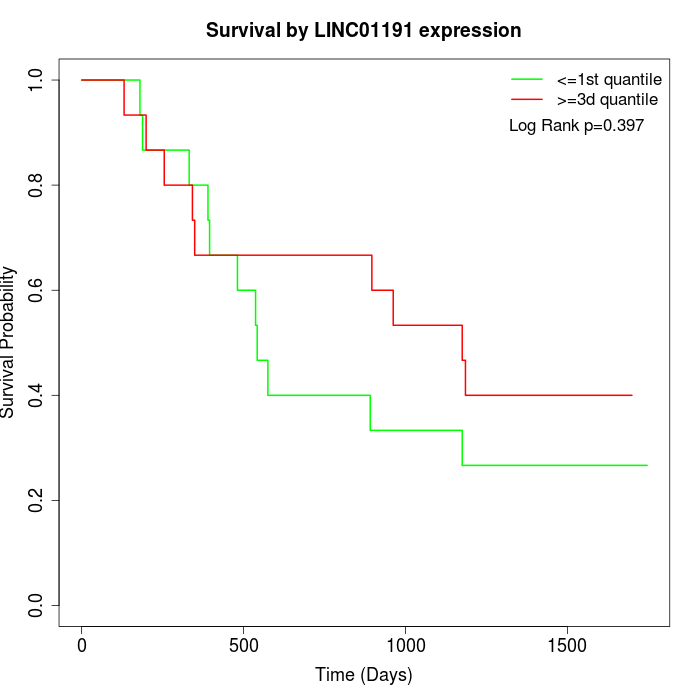
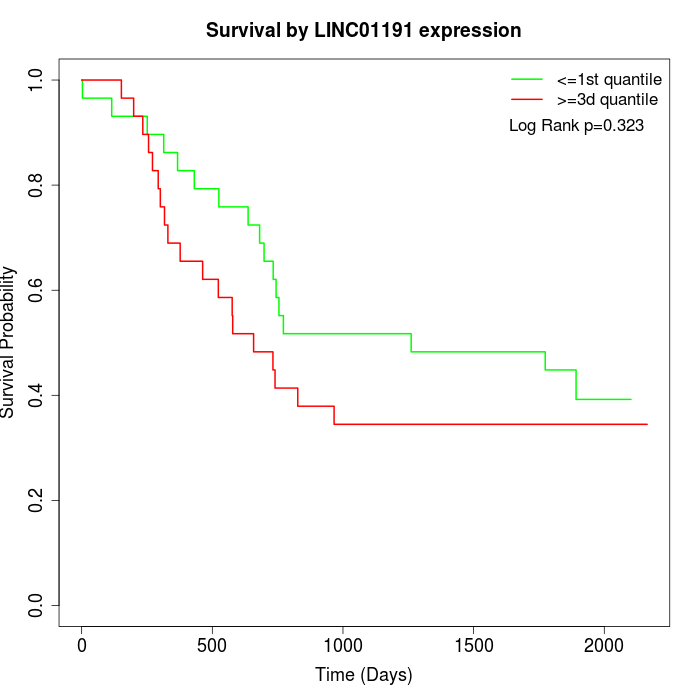
Survival by LINC01191 expression:
Note: Click image to view full size file.
Copy number change of LINC01191:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC01191 | 440900 | 6 | 2 | 22 | |
| GSE20123 | LINC01191 | 440900 | 6 | 2 | 22 | |
| GSE43470 | LINC01191 | 440900 | 3 | 1 | 39 | |
| GSE46452 | LINC01191 | 440900 | 1 | 4 | 54 | |
| GSE47630 | LINC01191 | 440900 | 6 | 0 | 34 | |
| GSE54993 | LINC01191 | 440900 | 0 | 6 | 64 | |
| GSE54994 | LINC01191 | 440900 | 11 | 0 | 42 | |
| GSE60625 | LINC01191 | 440900 | 0 | 3 | 8 | |
| GSE74703 | LINC01191 | 440900 | 3 | 1 | 32 | |
| GSE74704 | LINC01191 | 440900 | 2 | 2 | 16 |
Total number of gains: 38; Total number of losses: 21; Total Number of normals: 333.
Somatic mutations of LINC01191:
Generating mutation plots.
Highly correlated genes for LINC01191:
Showing top 20/54 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC01191 | BRSK1 | 0.658294 | 3 | 0 | 3 |
| LINC01191 | MYOG | 0.653005 | 3 | 0 | 3 |
| LINC01191 | TMEM238 | 0.651762 | 3 | 0 | 3 |
| LINC01191 | GRAP | 0.636657 | 3 | 0 | 3 |
| LINC01191 | NPAS1 | 0.635793 | 3 | 0 | 3 |
| LINC01191 | VCX2 | 0.629757 | 3 | 0 | 3 |
| LINC01191 | CRP | 0.628935 | 3 | 0 | 3 |
| LINC01191 | CT62 | 0.624732 | 3 | 0 | 3 |
| LINC01191 | FUT7 | 0.618811 | 4 | 0 | 4 |
| LINC01191 | CDH15 | 0.613902 | 3 | 0 | 3 |
| LINC01191 | KLK15 | 0.613256 | 3 | 0 | 3 |
| LINC01191 | CHST13 | 0.607359 | 4 | 0 | 3 |
| LINC01191 | GLTPD2 | 0.606209 | 4 | 0 | 3 |
| LINC01191 | VGF | 0.605306 | 4 | 0 | 4 |
| LINC01191 | CRYBA2 | 0.604527 | 3 | 0 | 3 |
| LINC01191 | BHLHE23 | 0.587961 | 3 | 0 | 3 |
| LINC01191 | TMEM151A | 0.586444 | 3 | 0 | 3 |
| LINC01191 | NSMF | 0.58419 | 4 | 0 | 3 |
| LINC01191 | RAP1GAP | 0.583535 | 3 | 0 | 3 |
| LINC01191 | CRYBB1 | 0.579498 | 5 | 0 | 4 |
For details and further investigation, click here