| Full name: VGF nerve growth factor inducible | Alias Symbol: SCG7|SgVII | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 7425 | HGNC ID: HGNC:12684 | Ensembl Gene: ENSG00000128564 | OMIM ID: 602186 |
| Drug and gene relationship at DGIdb | |||
Expression of VGF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | VGF | 7425 | 205586_x_at | 0.0537 | 0.8127 | |
| GSE20347 | VGF | 7425 | 205586_x_at | -0.1205 | 0.0867 | |
| GSE23400 | VGF | 7425 | 205586_x_at | -0.2457 | 0.0000 | |
| GSE26886 | VGF | 7425 | 205586_x_at | 0.2458 | 0.0250 | |
| GSE29001 | VGF | 7425 | 205586_x_at | -0.2337 | 0.1417 | |
| GSE38129 | VGF | 7425 | 205586_x_at | -0.0419 | 0.6880 | |
| GSE45670 | VGF | 7425 | 205586_x_at | 0.1063 | 0.3368 | |
| GSE53622 | VGF | 7425 | 121308 | 0.7357 | 0.0122 | |
| GSE53624 | VGF | 7425 | 121308 | 2.2399 | 0.0000 | |
| GSE63941 | VGF | 7425 | 205586_x_at | -0.0412 | 0.8675 | |
| GSE77861 | VGF | 7425 | 205586_x_at | -0.1252 | 0.3624 | |
| SRP219564 | VGF | 7425 | RNAseq | 1.7253 | 0.1068 | |
| TCGA | VGF | 7425 | RNAseq | -0.6638 | 0.0205 |
Upregulated datasets: 1; Downregulated datasets: 0.
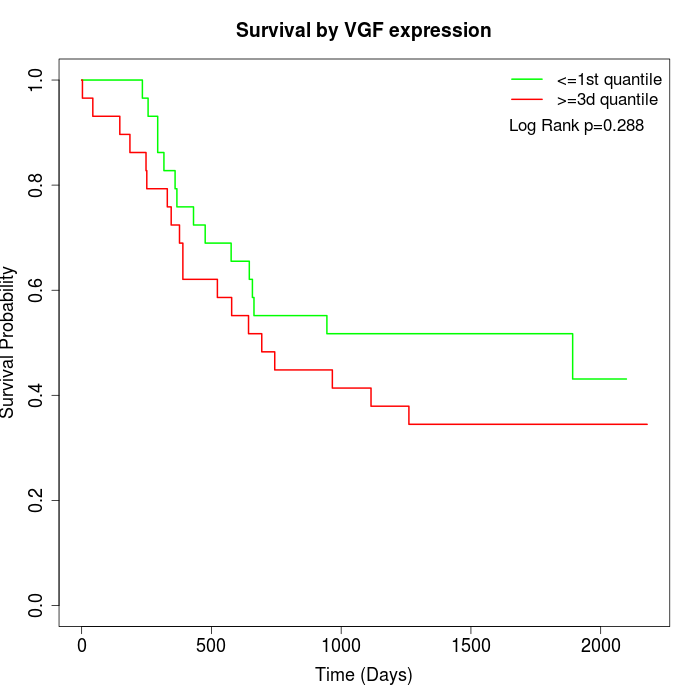
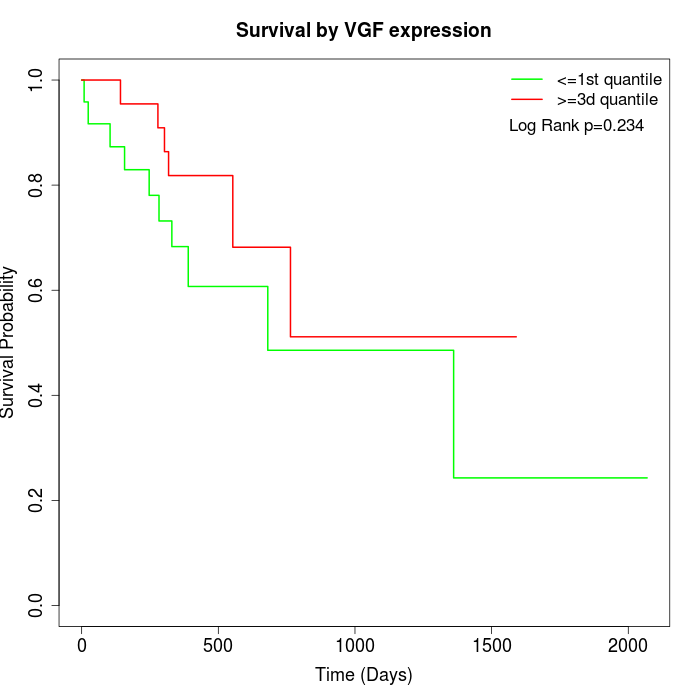
Survival by VGF expression:
Note: Click image to view full size file.
Copy number change of VGF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | VGF | 7425 | 13 | 0 | 17 | |
| GSE20123 | VGF | 7425 | 13 | 0 | 17 | |
| GSE43470 | VGF | 7425 | 7 | 2 | 34 | |
| GSE46452 | VGF | 7425 | 11 | 1 | 47 | |
| GSE47630 | VGF | 7425 | 7 | 3 | 30 | |
| GSE54993 | VGF | 7425 | 1 | 10 | 59 | |
| GSE54994 | VGF | 7425 | 15 | 3 | 35 | |
| GSE60625 | VGF | 7425 | 0 | 0 | 11 | |
| GSE74703 | VGF | 7425 | 7 | 1 | 28 | |
| GSE74704 | VGF | 7425 | 9 | 0 | 11 | |
| TCGA | VGF | 7425 | 53 | 6 | 37 |
Total number of gains: 136; Total number of losses: 26; Total Number of normals: 326.
Somatic mutations of VGF:
Generating mutation plots.
Highly correlated genes for VGF:
Showing top 20/619 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| VGF | KLK14 | 0.790552 | 3 | 0 | 3 |
| VGF | KAAG1 | 0.783099 | 3 | 0 | 3 |
| VGF | LMAN1L | 0.756333 | 3 | 0 | 3 |
| VGF | BFSP2 | 0.750406 | 3 | 0 | 3 |
| VGF | CEACAM21 | 0.748253 | 3 | 0 | 3 |
| VGF | CRP | 0.743243 | 4 | 0 | 4 |
| VGF | AVPR1A | 0.733818 | 3 | 0 | 3 |
| VGF | PGK2 | 0.726288 | 3 | 0 | 3 |
| VGF | CCL24 | 0.722819 | 3 | 0 | 3 |
| VGF | PKLR | 0.718337 | 5 | 0 | 5 |
| VGF | TPSD1 | 0.714637 | 4 | 0 | 4 |
| VGF | GRIN2C | 0.711139 | 4 | 0 | 4 |
| VGF | NPTXR | 0.705696 | 4 | 0 | 4 |
| VGF | GRIN2B | 0.702773 | 4 | 0 | 4 |
| VGF | INSRR | 0.69999 | 3 | 0 | 3 |
| VGF | OPRK1 | 0.697444 | 4 | 0 | 4 |
| VGF | GUCA1B | 0.697126 | 4 | 0 | 3 |
| VGF | CSH1 | 0.694072 | 3 | 0 | 3 |
| VGF | HCN4 | 0.691993 | 3 | 0 | 3 |
| VGF | CACNA1A | 0.68971 | 3 | 0 | 3 |
For details and further investigation, click here