| Full name: leucine rich repeats and transmembrane domains 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.33 | ||
| Entrez ID: 654429 | HGNC ID: HGNC:32443 | Ensembl Gene: ENSG00000166159 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LRTM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LRTM2 | 654429 | 1558530_at | -0.0526 | 0.8271 | |
| GSE26886 | LRTM2 | 654429 | 1558530_at | 0.0207 | 0.9243 | |
| GSE45670 | LRTM2 | 654429 | 1558530_at | -0.0357 | 0.7617 | |
| GSE53622 | LRTM2 | 654429 | 56761 | 0.0475 | 0.4980 | |
| GSE53624 | LRTM2 | 654429 | 56761 | 0.1171 | 0.1066 | |
| GSE63941 | LRTM2 | 654429 | 1558530_at | -0.0605 | 0.7687 | |
| GSE77861 | LRTM2 | 654429 | 1558530_at | -0.2189 | 0.1752 | |
| GSE97050 | LRTM2 | 654429 | A_23_P318616 | -0.0314 | 0.8841 | |
| TCGA | LRTM2 | 654429 | RNAseq | -0.7142 | 0.6460 |
Upregulated datasets: 0; Downregulated datasets: 0.
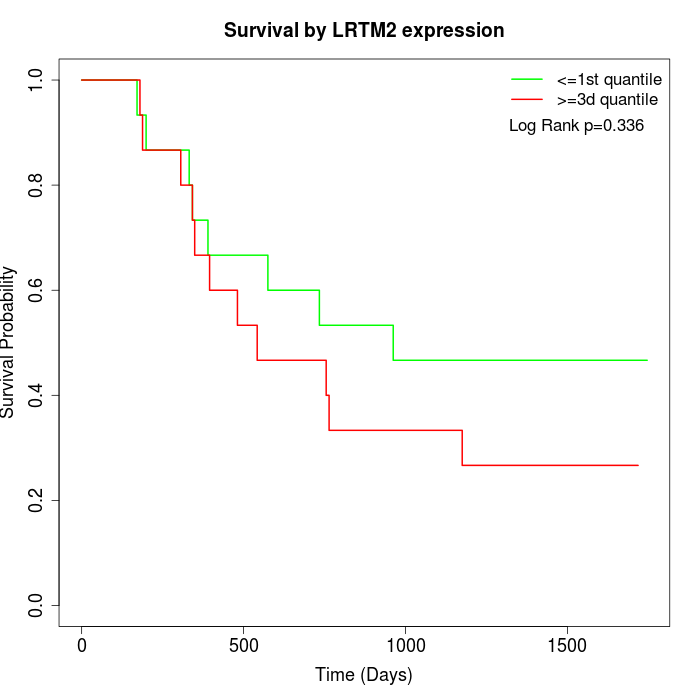
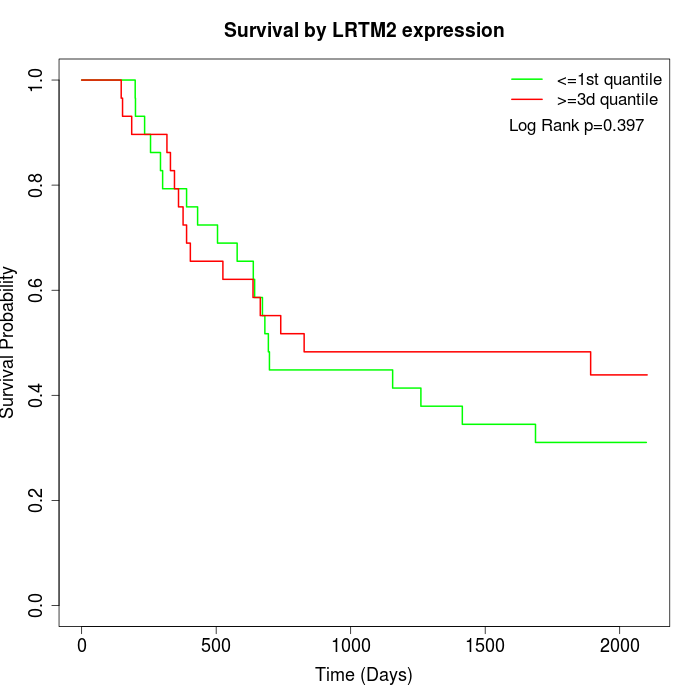
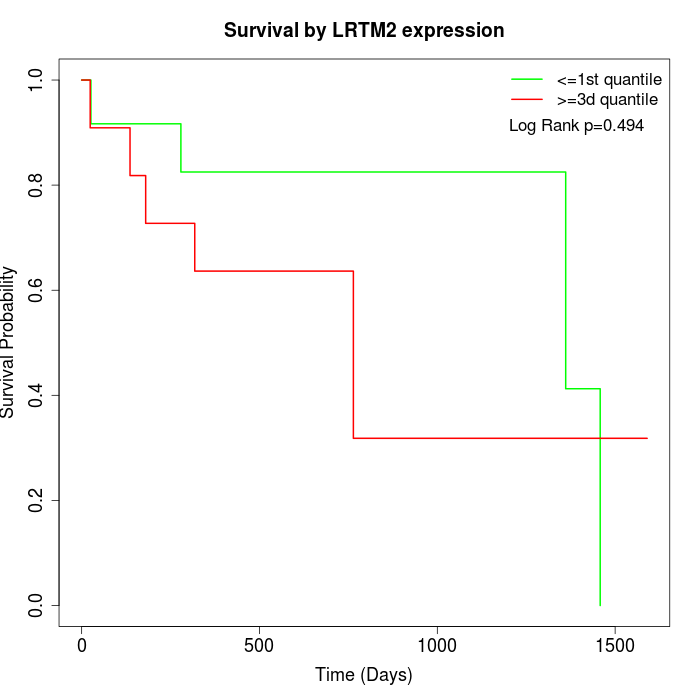
Survival by LRTM2 expression:
Note: Click image to view full size file.
Copy number change of LRTM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LRTM2 | 654429 | 6 | 4 | 20 | |
| GSE20123 | LRTM2 | 654429 | 6 | 4 | 20 | |
| GSE43470 | LRTM2 | 654429 | 11 | 3 | 29 | |
| GSE46452 | LRTM2 | 654429 | 10 | 1 | 48 | |
| GSE47630 | LRTM2 | 654429 | 12 | 2 | 26 | |
| GSE54993 | LRTM2 | 654429 | 1 | 10 | 59 | |
| GSE54994 | LRTM2 | 654429 | 10 | 2 | 41 | |
| GSE60625 | LRTM2 | 654429 | 0 | 1 | 10 | |
| GSE74703 | LRTM2 | 654429 | 10 | 2 | 24 | |
| GSE74704 | LRTM2 | 654429 | 4 | 2 | 14 | |
| TCGA | LRTM2 | 654429 | 40 | 6 | 50 |
Total number of gains: 110; Total number of losses: 37; Total Number of normals: 341.
Somatic mutations of LRTM2:
Generating mutation plots.
Highly correlated genes for LRTM2:
Showing top 20/318 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LRTM2 | FAM110B | 0.751608 | 3 | 0 | 3 |
| LRTM2 | TMPRSS5 | 0.74437 | 3 | 0 | 3 |
| LRTM2 | VRTN | 0.737586 | 3 | 0 | 3 |
| LRTM2 | OSM | 0.718756 | 3 | 0 | 3 |
| LRTM2 | BSX | 0.716754 | 3 | 0 | 3 |
| LRTM2 | SLC2A6 | 0.715961 | 3 | 0 | 3 |
| LRTM2 | SLC6A13 | 0.706504 | 3 | 0 | 3 |
| LRTM2 | MYO1A | 0.703914 | 4 | 0 | 4 |
| LRTM2 | MMD2 | 0.699639 | 4 | 0 | 4 |
| LRTM2 | NCR1 | 0.698593 | 3 | 0 | 3 |
| LRTM2 | SSPO | 0.695091 | 3 | 0 | 3 |
| LRTM2 | ITIH3 | 0.690732 | 4 | 0 | 4 |
| LRTM2 | GRIK4 | 0.688595 | 3 | 0 | 3 |
| LRTM2 | CFC1 | 0.686928 | 3 | 0 | 3 |
| LRTM2 | TSPEAR | 0.686778 | 3 | 0 | 3 |
| LRTM2 | CCL27 | 0.685342 | 4 | 0 | 3 |
| LRTM2 | C1QL2 | 0.684027 | 3 | 0 | 3 |
| LRTM2 | OR52L1 | 0.682391 | 3 | 0 | 3 |
| LRTM2 | TM6SF2 | 0.682157 | 3 | 0 | 3 |
| LRTM2 | CD28 | 0.680009 | 3 | 0 | 3 |
For details and further investigation, click here