| Full name: lysozyme g1 | Alias Symbol: SALW1939 | ||
| Type: protein-coding gene | Cytoband: 2q11.2 | ||
| Entrez ID: 129530 | HGNC ID: HGNC:27014 | Ensembl Gene: ENSG00000144214 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LYG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LYG1 | 129530 | 236706_at | 0.0003 | 0.9994 | |
| GSE26886 | LYG1 | 129530 | 236706_at | 0.3406 | 0.0723 | |
| GSE45670 | LYG1 | 129530 | 236706_at | 0.4415 | 0.0081 | |
| GSE53622 | LYG1 | 129530 | 53482 | 0.3188 | 0.0005 | |
| GSE53624 | LYG1 | 129530 | 53482 | 0.6879 | 0.0000 | |
| GSE63941 | LYG1 | 129530 | 236706_at | -0.0537 | 0.7915 | |
| GSE77861 | LYG1 | 129530 | 236706_at | 0.0369 | 0.8483 | |
| GSE97050 | LYG1 | 129530 | A_23_P165707 | 0.0774 | 0.6880 | |
| SRP064894 | LYG1 | 129530 | RNAseq | 1.0873 | 0.0000 | |
| TCGA | LYG1 | 129530 | RNAseq | 0.3434 | 0.3396 |
Upregulated datasets: 1; Downregulated datasets: 0.
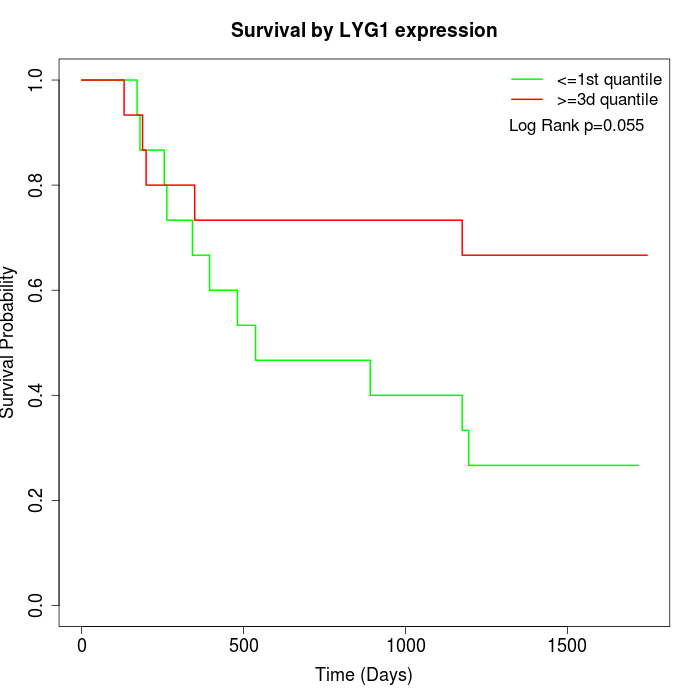
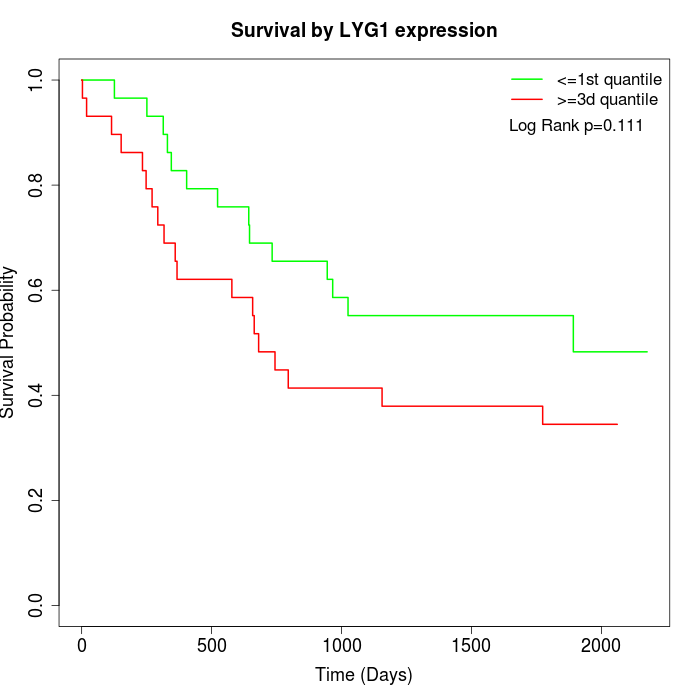
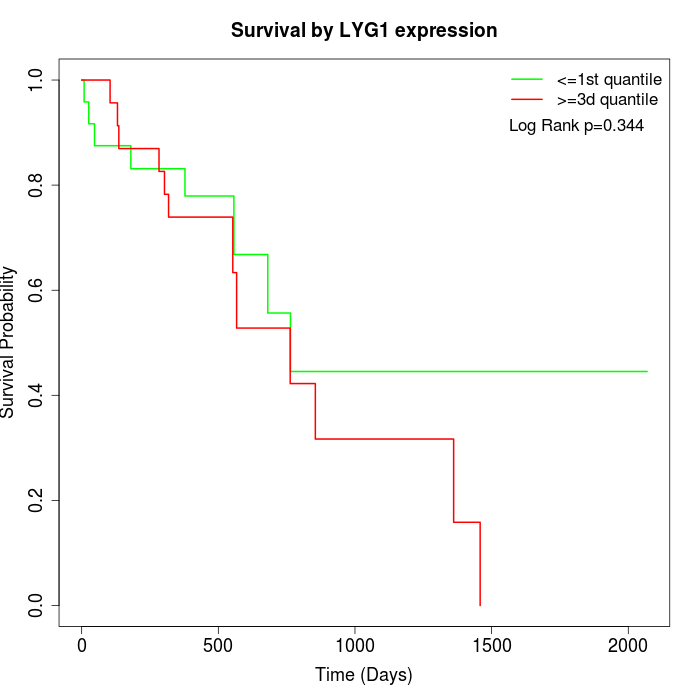
Survival by LYG1 expression:
Note: Click image to view full size file.
Copy number change of LYG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LYG1 | 129530 | 6 | 2 | 22 | |
| GSE20123 | LYG1 | 129530 | 6 | 2 | 22 | |
| GSE43470 | LYG1 | 129530 | 3 | 1 | 39 | |
| GSE46452 | LYG1 | 129530 | 2 | 3 | 54 | |
| GSE47630 | LYG1 | 129530 | 7 | 0 | 33 | |
| GSE54993 | LYG1 | 129530 | 0 | 6 | 64 | |
| GSE54994 | LYG1 | 129530 | 10 | 0 | 43 | |
| GSE60625 | LYG1 | 129530 | 0 | 3 | 8 | |
| GSE74703 | LYG1 | 129530 | 3 | 1 | 32 | |
| GSE74704 | LYG1 | 129530 | 4 | 1 | 15 | |
| TCGA | LYG1 | 129530 | 34 | 4 | 58 |
Total number of gains: 75; Total number of losses: 23; Total Number of normals: 390.
Somatic mutations of LYG1:
Generating mutation plots.
Highly correlated genes for LYG1:
Showing top 20/70 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LYG1 | ZNF527 | 0.748605 | 4 | 0 | 4 |
| LYG1 | ZNF444 | 0.746998 | 3 | 0 | 3 |
| LYG1 | ADD2 | 0.746022 | 3 | 0 | 3 |
| LYG1 | SCN5A | 0.742105 | 3 | 0 | 3 |
| LYG1 | CARD8 | 0.741529 | 3 | 0 | 3 |
| LYG1 | PNMA6A | 0.726732 | 3 | 0 | 3 |
| LYG1 | PCSK1N | 0.709305 | 4 | 0 | 3 |
| LYG1 | MSR1 | 0.68977 | 3 | 0 | 3 |
| LYG1 | ASMTL | 0.6865 | 4 | 0 | 3 |
| LYG1 | ATF7IP2 | 0.685588 | 3 | 0 | 3 |
| LYG1 | NR2E1 | 0.685576 | 3 | 0 | 3 |
| LYG1 | REM2 | 0.68405 | 3 | 0 | 3 |
| LYG1 | SLC27A1 | 0.683279 | 3 | 0 | 3 |
| LYG1 | TRIM17 | 0.680784 | 3 | 0 | 3 |
| LYG1 | ZDHHC22 | 0.672263 | 3 | 0 | 3 |
| LYG1 | SIGIRR | 0.671518 | 4 | 0 | 3 |
| LYG1 | JAKMIP3 | 0.669148 | 3 | 0 | 3 |
| LYG1 | TBC1D1 | 0.667918 | 3 | 0 | 3 |
| LYG1 | ACSF3 | 0.667848 | 3 | 0 | 3 |
| LYG1 | PHF21B | 0.666561 | 4 | 0 | 3 |
For details and further investigation, click here