| Full name: RRAD and GEM like GTPase 2 | Alias Symbol: FLJ38964 | ||
| Type: protein-coding gene | Cytoband: 14q11.2 | ||
| Entrez ID: 161253 | HGNC ID: HGNC:20248 | Ensembl Gene: ENSG00000139890 | OMIM ID: 616955 |
| Drug and gene relationship at DGIdb | |||
Expression of REM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | REM2 | 161253 | 235699_at | 0.2619 | 0.4595 | |
| GSE26886 | REM2 | 161253 | 235699_at | 0.2457 | 0.1033 | |
| GSE45670 | REM2 | 161253 | 235699_at | 0.1522 | 0.1807 | |
| GSE53622 | REM2 | 161253 | 104756 | 0.0923 | 0.2628 | |
| GSE53624 | REM2 | 161253 | 104756 | 0.2078 | 0.0062 | |
| GSE63941 | REM2 | 161253 | 235699_at | 0.7793 | 0.0030 | |
| GSE77861 | REM2 | 161253 | 235699_at | 0.0145 | 0.8885 | |
| GSE97050 | REM2 | 161253 | A_33_P3305487 | 0.1069 | 0.5608 | |
| SRP133303 | REM2 | 161253 | RNAseq | -0.2435 | 0.3309 | |
| SRP159526 | REM2 | 161253 | RNAseq | 0.2290 | 0.6971 | |
| SRP219564 | REM2 | 161253 | RNAseq | 0.4336 | 0.3750 | |
| TCGA | REM2 | 161253 | RNAseq | 0.5796 | 0.0197 |
Upregulated datasets: 0; Downregulated datasets: 0.
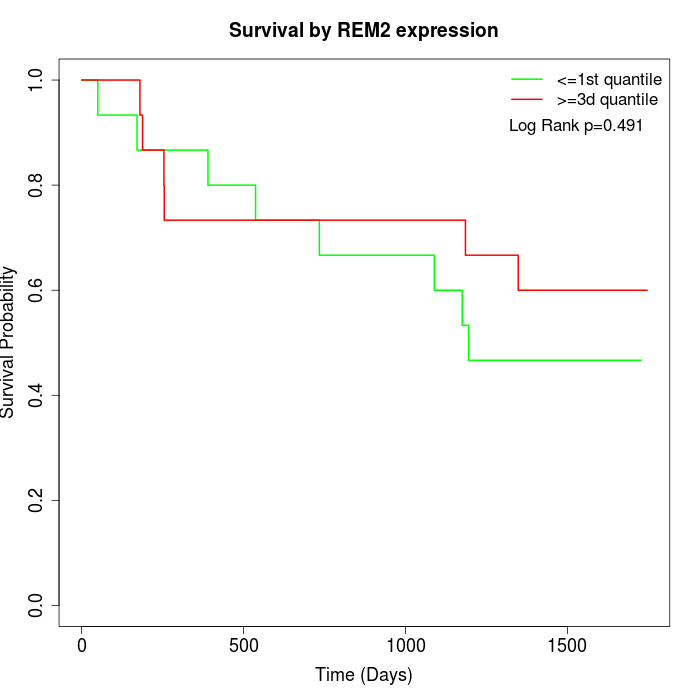
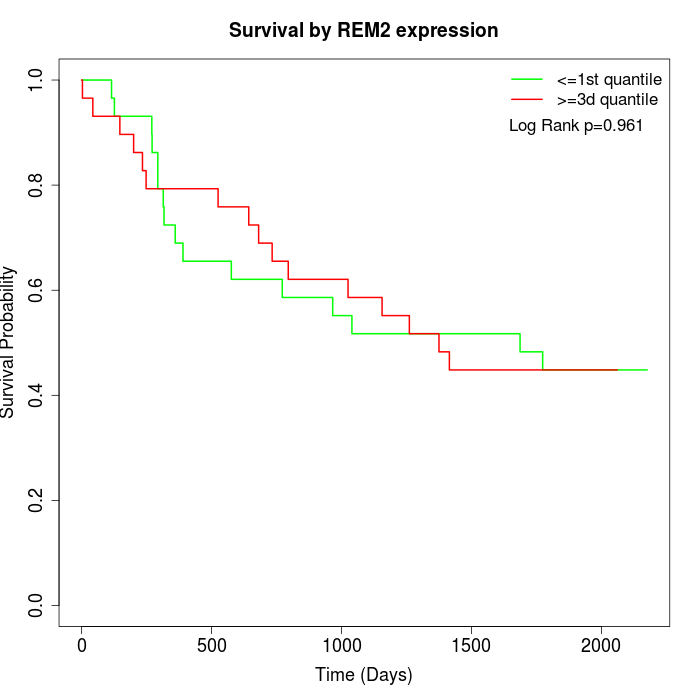
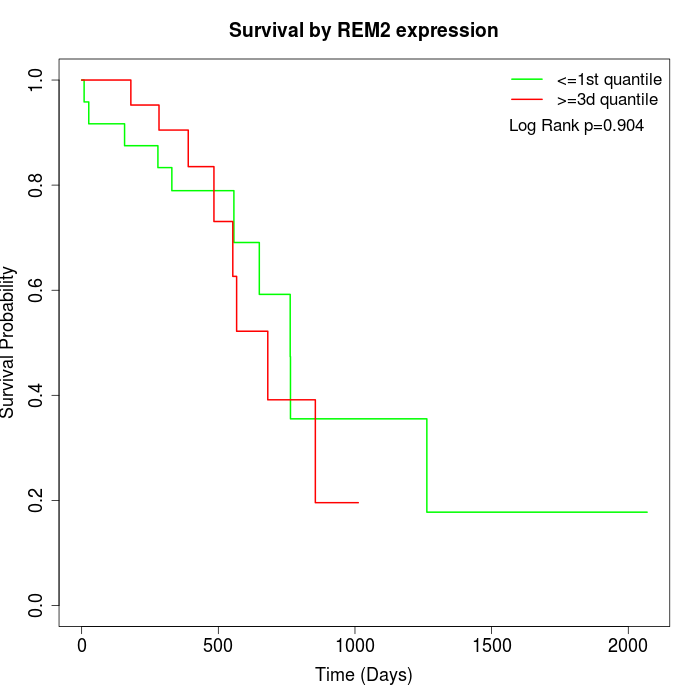
Survival by REM2 expression:
Note: Click image to view full size file.
Copy number change of REM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | REM2 | 161253 | 9 | 2 | 19 | |
| GSE20123 | REM2 | 161253 | 9 | 2 | 19 | |
| GSE43470 | REM2 | 161253 | 7 | 2 | 34 | |
| GSE46452 | REM2 | 161253 | 17 | 2 | 40 | |
| GSE47630 | REM2 | 161253 | 11 | 10 | 19 | |
| GSE54993 | REM2 | 161253 | 3 | 11 | 56 | |
| GSE54994 | REM2 | 161253 | 18 | 5 | 30 | |
| GSE60625 | REM2 | 161253 | 0 | 2 | 9 | |
| GSE74703 | REM2 | 161253 | 6 | 2 | 28 | |
| GSE74704 | REM2 | 161253 | 4 | 1 | 15 | |
| TCGA | REM2 | 161253 | 26 | 16 | 54 |
Total number of gains: 110; Total number of losses: 55; Total Number of normals: 323.
Somatic mutations of REM2:
Generating mutation plots.
Highly correlated genes for REM2:
Showing top 20/110 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| REM2 | ZMYND15 | 0.750512 | 3 | 0 | 3 |
| REM2 | LAS1L | 0.742075 | 3 | 0 | 3 |
| REM2 | HK3 | 0.734827 | 3 | 0 | 3 |
| REM2 | NECAB3 | 0.730288 | 3 | 0 | 3 |
| REM2 | PRR18 | 0.72932 | 3 | 0 | 3 |
| REM2 | PHF21A | 0.72639 | 3 | 0 | 3 |
| REM2 | PCSK1N | 0.726319 | 4 | 0 | 3 |
| REM2 | IL27RA | 0.721483 | 3 | 0 | 3 |
| REM2 | KRT9 | 0.710568 | 3 | 0 | 3 |
| REM2 | NOXA1 | 0.702292 | 3 | 0 | 3 |
| REM2 | ASMTL | 0.700728 | 3 | 0 | 3 |
| REM2 | GPN2 | 0.698801 | 3 | 0 | 3 |
| REM2 | SHISA8 | 0.695136 | 3 | 0 | 3 |
| REM2 | CPAMD8 | 0.690531 | 5 | 0 | 4 |
| REM2 | CPNE7 | 0.689341 | 3 | 0 | 3 |
| REM2 | LYG1 | 0.68405 | 3 | 0 | 3 |
| REM2 | UBXN2B | 0.677784 | 4 | 0 | 3 |
| REM2 | LAMA3 | 0.675248 | 3 | 0 | 3 |
| REM2 | INF2 | 0.672188 | 3 | 0 | 3 |
| REM2 | OR7A17 | 0.671331 | 3 | 0 | 3 |
For details and further investigation, click here