| Full name: MALT1 paracaspase | Alias Symbol: PCASP1 | ||
| Type: protein-coding gene | Cytoband: 18q21.32 | ||
| Entrez ID: 10892 | HGNC ID: HGNC:6819 | Ensembl Gene: ENSG00000172175 | OMIM ID: 604860 |
| Drug and gene relationship at DGIdb | |||
MALT1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04660 | T cell receptor signaling pathway | |
| hsa04662 | B cell receptor signaling pathway | |
| hsa05152 | Tuberculosis |
Expression of MALT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MALT1 | 10892 | 210018_x_at | -0.2032 | 0.8656 | |
| GSE20347 | MALT1 | 10892 | 208309_s_at | -0.3010 | 0.1906 | |
| GSE23400 | MALT1 | 10892 | 210018_x_at | -0.1695 | 0.1061 | |
| GSE26886 | MALT1 | 10892 | 210018_x_at | -0.6867 | 0.0098 | |
| GSE29001 | MALT1 | 10892 | 210017_at | -0.5978 | 0.1911 | |
| GSE38129 | MALT1 | 10892 | 210018_x_at | 0.0657 | 0.7664 | |
| GSE45670 | MALT1 | 10892 | 210018_x_at | 0.0088 | 0.9715 | |
| GSE53622 | MALT1 | 10892 | 98375 | -0.1823 | 0.0310 | |
| GSE53624 | MALT1 | 10892 | 98375 | -0.4854 | 0.0000 | |
| GSE63941 | MALT1 | 10892 | 210018_x_at | -0.3620 | 0.6452 | |
| GSE77861 | MALT1 | 10892 | 210018_x_at | -0.3322 | 0.2012 | |
| GSE97050 | MALT1 | 10892 | A_24_P116909 | 0.1107 | 0.7040 | |
| SRP007169 | MALT1 | 10892 | RNAseq | -0.9538 | 0.0061 | |
| SRP008496 | MALT1 | 10892 | RNAseq | -0.6752 | 0.0172 | |
| SRP064894 | MALT1 | 10892 | RNAseq | -0.4317 | 0.0582 | |
| SRP133303 | MALT1 | 10892 | RNAseq | -0.1775 | 0.3541 | |
| SRP159526 | MALT1 | 10892 | RNAseq | -0.5845 | 0.0498 | |
| SRP193095 | MALT1 | 10892 | RNAseq | -0.3399 | 0.0256 | |
| SRP219564 | MALT1 | 10892 | RNAseq | -0.9193 | 0.0434 | |
| TCGA | MALT1 | 10892 | RNAseq | 0.1699 | 0.0066 |
Upregulated datasets: 0; Downregulated datasets: 0.
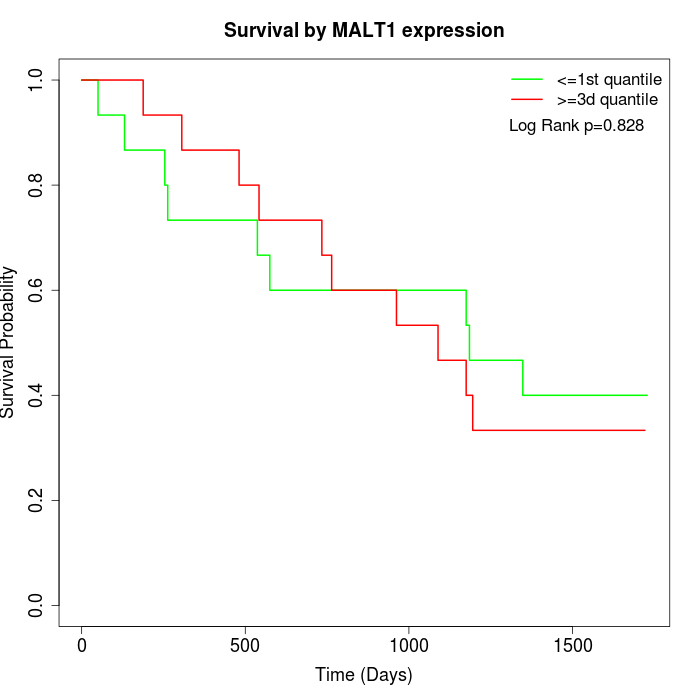
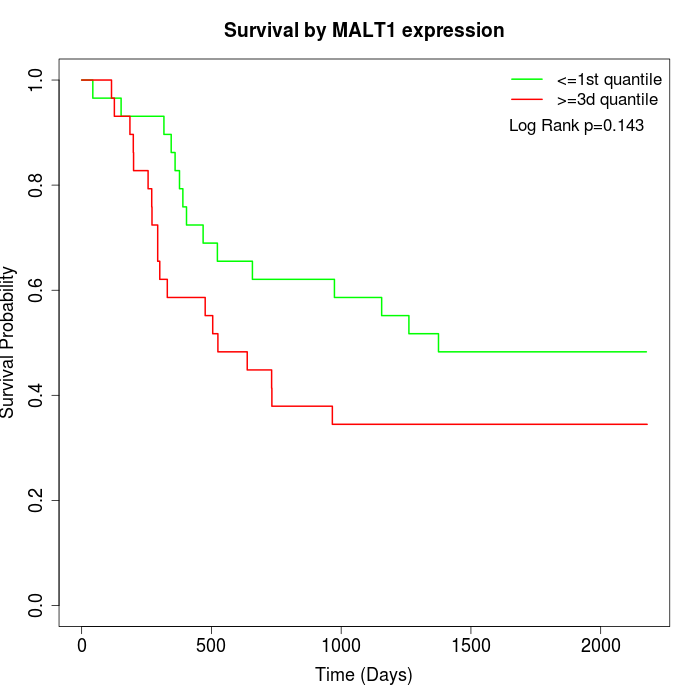
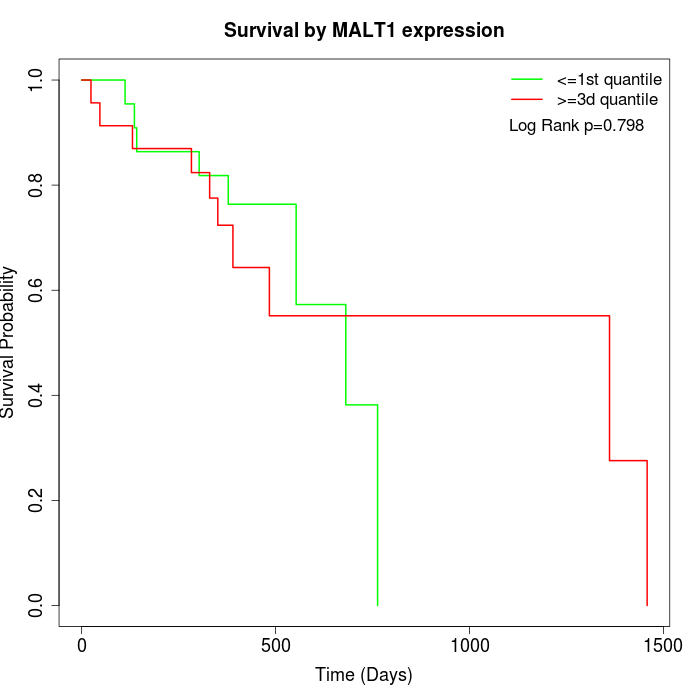
Survival by MALT1 expression:
Note: Click image to view full size file.
Copy number change of MALT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MALT1 | 10892 | 2 | 8 | 20 | |
| GSE20123 | MALT1 | 10892 | 2 | 8 | 20 | |
| GSE43470 | MALT1 | 10892 | 0 | 5 | 38 | |
| GSE46452 | MALT1 | 10892 | 1 | 26 | 32 | |
| GSE47630 | MALT1 | 10892 | 5 | 20 | 15 | |
| GSE54993 | MALT1 | 10892 | 10 | 0 | 60 | |
| GSE54994 | MALT1 | 10892 | 0 | 18 | 35 | |
| GSE60625 | MALT1 | 10892 | 0 | 4 | 7 | |
| GSE74703 | MALT1 | 10892 | 0 | 5 | 31 | |
| GSE74704 | MALT1 | 10892 | 2 | 5 | 13 | |
| TCGA | MALT1 | 10892 | 9 | 44 | 43 |
Total number of gains: 31; Total number of losses: 143; Total Number of normals: 314.
Somatic mutations of MALT1:
Generating mutation plots.
Highly correlated genes for MALT1:
Showing top 20/605 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MALT1 | EXOC6B | 0.773929 | 3 | 0 | 3 |
| MALT1 | THAP2 | 0.748292 | 4 | 0 | 4 |
| MALT1 | RAD23B | 0.747484 | 3 | 0 | 3 |
| MALT1 | CCDC25 | 0.738389 | 3 | 0 | 3 |
| MALT1 | ATXN7 | 0.732963 | 3 | 0 | 3 |
| MALT1 | SLC38A5 | 0.726989 | 3 | 0 | 3 |
| MALT1 | NDUFS4 | 0.723006 | 5 | 0 | 5 |
| MALT1 | NSFL1C | 0.719216 | 3 | 0 | 3 |
| MALT1 | TCHP | 0.718638 | 3 | 0 | 3 |
| MALT1 | CGRRF1 | 0.713909 | 3 | 0 | 3 |
| MALT1 | MAP3K8 | 0.709337 | 4 | 0 | 3 |
| MALT1 | ATP6V0D1 | 0.70609 | 5 | 0 | 5 |
| MALT1 | KLHL17 | 0.704856 | 3 | 0 | 3 |
| MALT1 | RNFT1 | 0.70421 | 4 | 0 | 4 |
| MALT1 | PHKB | 0.703819 | 4 | 0 | 4 |
| MALT1 | ZNF844 | 0.701594 | 3 | 0 | 3 |
| MALT1 | DPH5 | 0.700349 | 3 | 0 | 3 |
| MALT1 | MLKL | 0.699293 | 4 | 0 | 4 |
| MALT1 | ZNF143 | 0.697387 | 4 | 0 | 4 |
| MALT1 | SLC43A3 | 0.697259 | 4 | 0 | 3 |
For details and further investigation, click here