| Full name: mitogen-activated protein kinase kinase kinase 8 | Alias Symbol: Tpl-2|EST|c-COT|MEKK8 | ||
| Type: protein-coding gene | Cytoband: 10p11.23 | ||
| Entrez ID: 1326 | HGNC ID: HGNC:6860 | Ensembl Gene: ENSG00000107968 | OMIM ID: 191195 |
| Drug and gene relationship at DGIdb | |||
MAP3K8 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04620 | Toll-like receptor signaling pathway | |
| hsa04660 | T cell receptor signaling pathway | |
| hsa04668 | TNF signaling pathway |
Expression of MAP3K8:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP3K8 | 1326 | 205027_s_at | -0.4313 | 0.6371 | |
| GSE20347 | MAP3K8 | 1326 | 205027_s_at | -0.6147 | 0.0603 | |
| GSE23400 | MAP3K8 | 1326 | 205027_s_at | -0.0592 | 0.5049 | |
| GSE26886 | MAP3K8 | 1326 | 205027_s_at | -1.1424 | 0.0060 | |
| GSE29001 | MAP3K8 | 1326 | 205027_s_at | -0.8916 | 0.1853 | |
| GSE38129 | MAP3K8 | 1326 | 205027_s_at | -0.5701 | 0.0564 | |
| GSE45670 | MAP3K8 | 1326 | 205027_s_at | -0.0028 | 0.9925 | |
| GSE53622 | MAP3K8 | 1326 | 26031 | -0.4587 | 0.0001 | |
| GSE53624 | MAP3K8 | 1326 | 26031 | -0.5421 | 0.0000 | |
| GSE63941 | MAP3K8 | 1326 | 205027_s_at | -1.7849 | 0.0720 | |
| GSE77861 | MAP3K8 | 1326 | 235421_at | -0.0145 | 0.9735 | |
| GSE97050 | MAP3K8 | 1326 | A_33_P3246505 | -0.2505 | 0.3748 | |
| SRP007169 | MAP3K8 | 1326 | RNAseq | -0.8768 | 0.0756 | |
| SRP008496 | MAP3K8 | 1326 | RNAseq | -0.8981 | 0.0032 | |
| SRP064894 | MAP3K8 | 1326 | RNAseq | -0.7145 | 0.0286 | |
| SRP133303 | MAP3K8 | 1326 | RNAseq | -0.1527 | 0.6417 | |
| SRP159526 | MAP3K8 | 1326 | RNAseq | -0.9097 | 0.0450 | |
| SRP193095 | MAP3K8 | 1326 | RNAseq | 0.2105 | 0.2850 | |
| SRP219564 | MAP3K8 | 1326 | RNAseq | -0.4390 | 0.3822 | |
| TCGA | MAP3K8 | 1326 | RNAseq | -0.1308 | 0.2453 |
Upregulated datasets: 0; Downregulated datasets: 1.
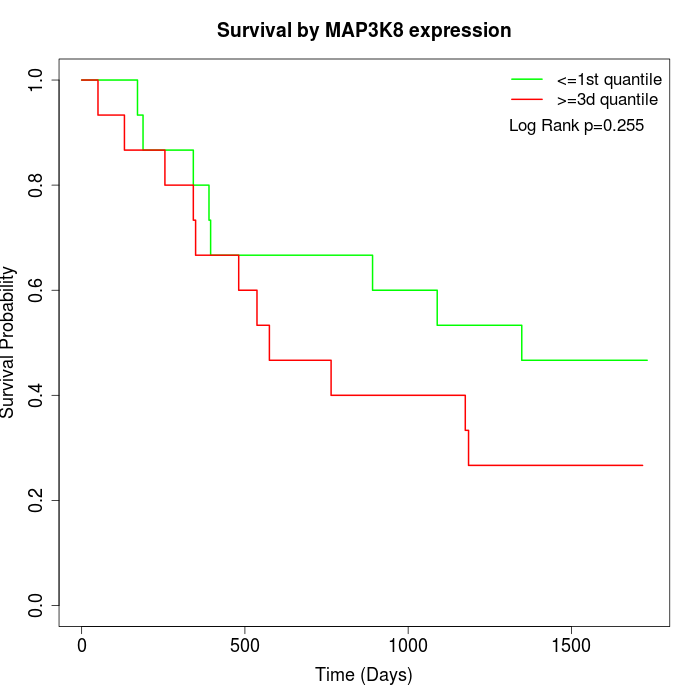
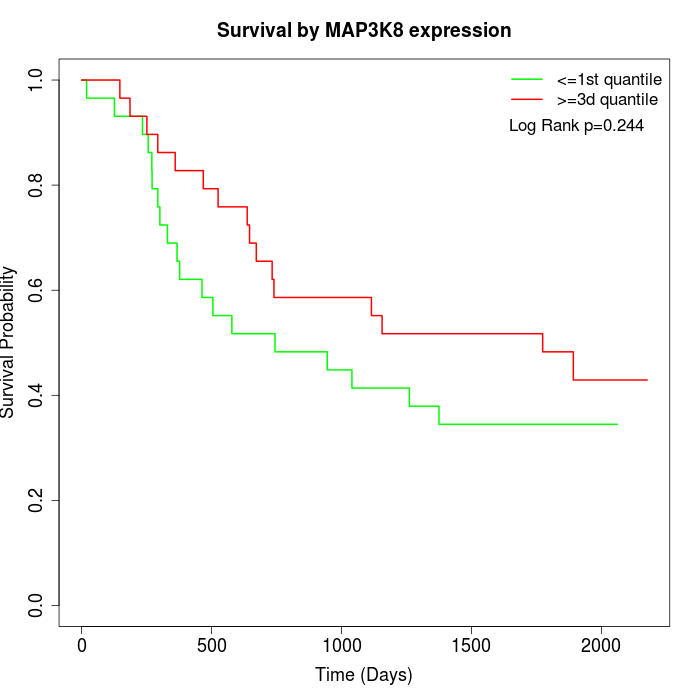
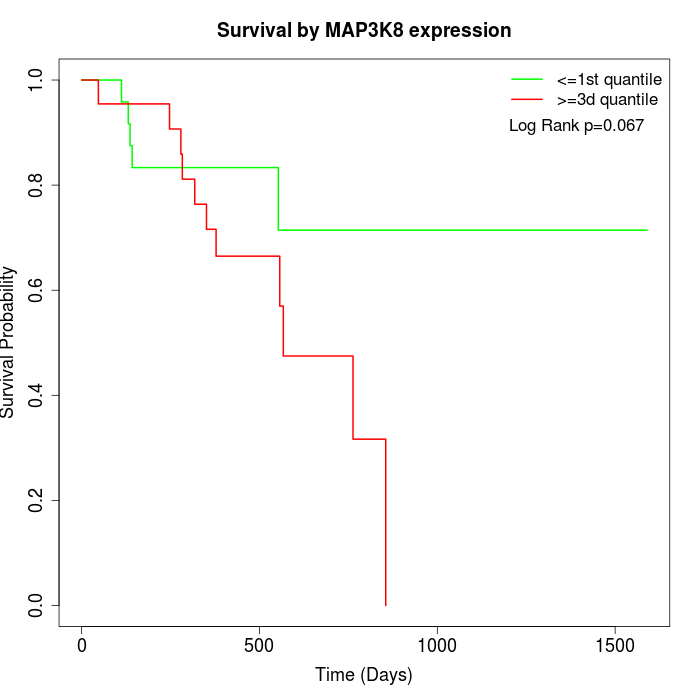
Survival by MAP3K8 expression:
Note: Click image to view full size file.
Copy number change of MAP3K8:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP3K8 | 1326 | 5 | 7 | 18 | |
| GSE20123 | MAP3K8 | 1326 | 5 | 6 | 19 | |
| GSE43470 | MAP3K8 | 1326 | 3 | 4 | 36 | |
| GSE46452 | MAP3K8 | 1326 | 1 | 14 | 44 | |
| GSE47630 | MAP3K8 | 1326 | 5 | 15 | 20 | |
| GSE54993 | MAP3K8 | 1326 | 9 | 0 | 61 | |
| GSE54994 | MAP3K8 | 1326 | 3 | 9 | 41 | |
| GSE60625 | MAP3K8 | 1326 | 0 | 0 | 11 | |
| GSE74703 | MAP3K8 | 1326 | 2 | 2 | 32 | |
| GSE74704 | MAP3K8 | 1326 | 0 | 5 | 15 | |
| TCGA | MAP3K8 | 1326 | 19 | 23 | 54 |
Total number of gains: 52; Total number of losses: 85; Total Number of normals: 351.
Somatic mutations of MAP3K8:
Generating mutation plots.
Highly correlated genes for MAP3K8:
Showing top 20/334 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP3K8 | ATP6V0D1 | 0.811864 | 3 | 0 | 3 |
| MAP3K8 | TMEM50A | 0.781671 | 3 | 0 | 3 |
| MAP3K8 | PPP4R2 | 0.778474 | 3 | 0 | 3 |
| MAP3K8 | RDX | 0.775832 | 3 | 0 | 3 |
| MAP3K8 | WARS2 | 0.764048 | 3 | 0 | 3 |
| MAP3K8 | NDN | 0.743113 | 3 | 0 | 3 |
| MAP3K8 | HSBP1 | 0.721741 | 3 | 0 | 3 |
| MAP3K8 | NPAT | 0.720461 | 3 | 0 | 3 |
| MAP3K8 | MALT1 | 0.709337 | 4 | 0 | 3 |
| MAP3K8 | C9orf72 | 0.709206 | 4 | 0 | 3 |
| MAP3K8 | FBXW11 | 0.706804 | 3 | 0 | 3 |
| MAP3K8 | TMX4 | 0.704625 | 3 | 0 | 3 |
| MAP3K8 | EPG5 | 0.697602 | 3 | 0 | 3 |
| MAP3K8 | PPP2R3C | 0.69305 | 3 | 0 | 3 |
| MAP3K8 | DNAJA2 | 0.688206 | 4 | 0 | 4 |
| MAP3K8 | TIFA | 0.687107 | 3 | 0 | 3 |
| MAP3K8 | MEGF9 | 0.686593 | 3 | 0 | 3 |
| MAP3K8 | JMJD1C | 0.68575 | 5 | 0 | 4 |
| MAP3K8 | SLC25A3 | 0.681003 | 3 | 0 | 3 |
| MAP3K8 | SEC24B | 0.680321 | 4 | 0 | 3 |
For details and further investigation, click here