| Full name: mitogen-activated protein kinase kinase 5 | Alias Symbol: MEK5|MAPKK5|HsT17454 | ||
| Type: protein-coding gene | Cytoband: 15q23 | ||
| Entrez ID: 5607 | HGNC ID: HGNC:6845 | Ensembl Gene: ENSG00000137764 | OMIM ID: 602520 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MAP2K5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04540 | Gap junction | |
| hsa04722 | Neurotrophin signaling pathway | |
| hsa04921 | Oxytocin signaling pathway |
Expression of MAP2K5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP2K5 | 5607 | 211370_s_at | -0.1750 | 0.8055 | |
| GSE20347 | MAP2K5 | 5607 | 204756_at | 0.0778 | 0.2480 | |
| GSE23400 | MAP2K5 | 5607 | 210482_x_at | -0.1131 | 0.0016 | |
| GSE26886 | MAP2K5 | 5607 | 204756_at | 0.4001 | 0.0005 | |
| GSE29001 | MAP2K5 | 5607 | 204756_at | 0.1925 | 0.3179 | |
| GSE38129 | MAP2K5 | 5607 | 204756_at | -0.0527 | 0.5066 | |
| GSE45670 | MAP2K5 | 5607 | 204756_at | -0.0710 | 0.4733 | |
| GSE53622 | MAP2K5 | 5607 | 82505 | 0.1425 | 0.0594 | |
| GSE53624 | MAP2K5 | 5607 | 82505 | 0.1190 | 0.2518 | |
| GSE63941 | MAP2K5 | 5607 | 204756_at | -0.2147 | 0.4901 | |
| GSE77861 | MAP2K5 | 5607 | 204756_at | -0.0239 | 0.9000 | |
| GSE97050 | MAP2K5 | 5607 | A_24_P356130 | -0.3409 | 0.2307 | |
| SRP007169 | MAP2K5 | 5607 | RNAseq | -0.0248 | 0.9564 | |
| SRP064894 | MAP2K5 | 5607 | RNAseq | 0.4350 | 0.0479 | |
| SRP133303 | MAP2K5 | 5607 | RNAseq | -0.1022 | 0.5216 | |
| SRP159526 | MAP2K5 | 5607 | RNAseq | 0.0177 | 0.9461 | |
| SRP193095 | MAP2K5 | 5607 | RNAseq | 0.1346 | 0.2183 | |
| SRP219564 | MAP2K5 | 5607 | RNAseq | 0.1298 | 0.7699 | |
| TCGA | MAP2K5 | 5607 | RNAseq | -0.1116 | 0.0551 |
Upregulated datasets: 0; Downregulated datasets: 0.
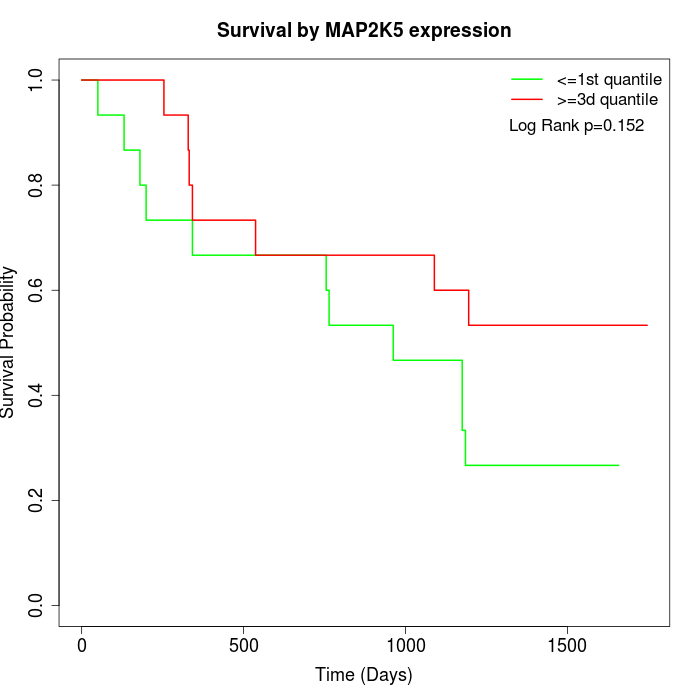
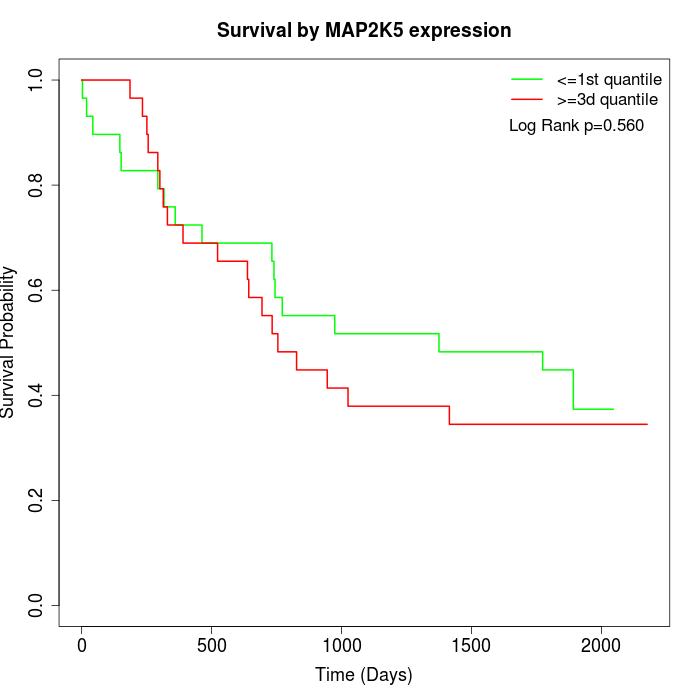
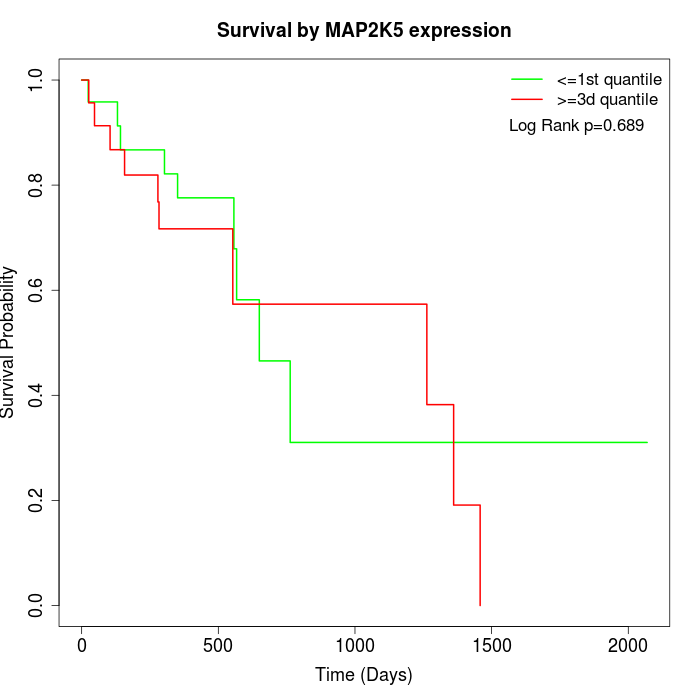
Survival by MAP2K5 expression:
Note: Click image to view full size file.
Copy number change of MAP2K5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP2K5 | 5607 | 7 | 1 | 22 | |
| GSE20123 | MAP2K5 | 5607 | 7 | 1 | 22 | |
| GSE43470 | MAP2K5 | 5607 | 4 | 6 | 33 | |
| GSE46452 | MAP2K5 | 5607 | 3 | 7 | 49 | |
| GSE47630 | MAP2K5 | 5607 | 8 | 10 | 22 | |
| GSE54993 | MAP2K5 | 5607 | 5 | 6 | 59 | |
| GSE54994 | MAP2K5 | 5607 | 6 | 7 | 40 | |
| GSE60625 | MAP2K5 | 5607 | 4 | 0 | 7 | |
| GSE74703 | MAP2K5 | 5607 | 4 | 3 | 29 | |
| GSE74704 | MAP2K5 | 5607 | 3 | 1 | 16 | |
| TCGA | MAP2K5 | 5607 | 10 | 16 | 70 |
Total number of gains: 61; Total number of losses: 58; Total Number of normals: 369.
Somatic mutations of MAP2K5:
Generating mutation plots.
Highly correlated genes for MAP2K5:
Showing top 20/357 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP2K5 | TMTC1 | 0.807292 | 3 | 0 | 3 |
| MAP2K5 | PTPRU | 0.795283 | 3 | 0 | 3 |
| MAP2K5 | MXRA7 | 0.784526 | 3 | 0 | 3 |
| MAP2K5 | PKIG | 0.782199 | 3 | 0 | 3 |
| MAP2K5 | STUB1 | 0.773007 | 3 | 0 | 3 |
| MAP2K5 | KLHL5 | 0.769053 | 3 | 0 | 3 |
| MAP2K5 | SRPX | 0.753923 | 3 | 0 | 3 |
| MAP2K5 | LRRC23 | 0.748299 | 4 | 0 | 3 |
| MAP2K5 | UHRF1BP1L | 0.747757 | 3 | 0 | 3 |
| MAP2K5 | RNF14 | 0.738544 | 3 | 0 | 3 |
| MAP2K5 | CMC1 | 0.735904 | 3 | 0 | 3 |
| MAP2K5 | C20orf27 | 0.735842 | 3 | 0 | 3 |
| MAP2K5 | MACF1 | 0.724703 | 3 | 0 | 3 |
| MAP2K5 | FHOD3 | 0.720673 | 3 | 0 | 3 |
| MAP2K5 | ARL2 | 0.719458 | 4 | 0 | 4 |
| MAP2K5 | KLHL13 | 0.718485 | 4 | 0 | 3 |
| MAP2K5 | JAM3 | 0.717544 | 3 | 0 | 3 |
| MAP2K5 | TSHZ3 | 0.717446 | 3 | 0 | 3 |
| MAP2K5 | RRP9 | 0.715156 | 3 | 0 | 3 |
| MAP2K5 | DR1 | 0.708221 | 3 | 0 | 3 |
For details and further investigation, click here