| Full name: microtubule associated protein 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 4134 | HGNC ID: HGNC:6862 | Ensembl Gene: ENSG00000047849 | OMIM ID: 157132 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP4 | 4134 | 212566_at | -0.1843 | 0.6892 | |
| GSE20347 | MAP4 | 4134 | 243_g_at | -0.3255 | 0.0576 | |
| GSE23400 | MAP4 | 4134 | 212566_at | -0.1594 | 0.0925 | |
| GSE26886 | MAP4 | 4134 | 212566_at | 0.3242 | 0.1250 | |
| GSE29001 | MAP4 | 4134 | 212566_at | -0.0070 | 0.9871 | |
| GSE38129 | MAP4 | 4134 | 212566_at | -0.6951 | 0.0097 | |
| GSE45670 | MAP4 | 4134 | 243_g_at | 0.2241 | 0.1985 | |
| GSE53622 | MAP4 | 4134 | 52470 | 0.0019 | 0.9722 | |
| GSE53624 | MAP4 | 4134 | 52470 | 0.0227 | 0.7536 | |
| GSE63941 | MAP4 | 4134 | 243_g_at | -2.1427 | 0.0002 | |
| GSE77861 | MAP4 | 4134 | 243_g_at | 0.0068 | 0.9833 | |
| GSE97050 | MAP4 | 4134 | A_33_P3354137 | 0.0695 | 0.7690 | |
| SRP007169 | MAP4 | 4134 | RNAseq | 0.0908 | 0.7948 | |
| SRP008496 | MAP4 | 4134 | RNAseq | 0.2132 | 0.4052 | |
| SRP064894 | MAP4 | 4134 | RNAseq | 0.2273 | 0.1355 | |
| SRP133303 | MAP4 | 4134 | RNAseq | 0.1422 | 0.3486 | |
| SRP159526 | MAP4 | 4134 | RNAseq | -0.6140 | 0.0344 | |
| SRP193095 | MAP4 | 4134 | RNAseq | 0.1047 | 0.3689 | |
| SRP219564 | MAP4 | 4134 | RNAseq | -0.3539 | 0.4445 | |
| TCGA | MAP4 | 4134 | RNAseq | -0.0983 | 0.0432 |
Upregulated datasets: 0; Downregulated datasets: 1.
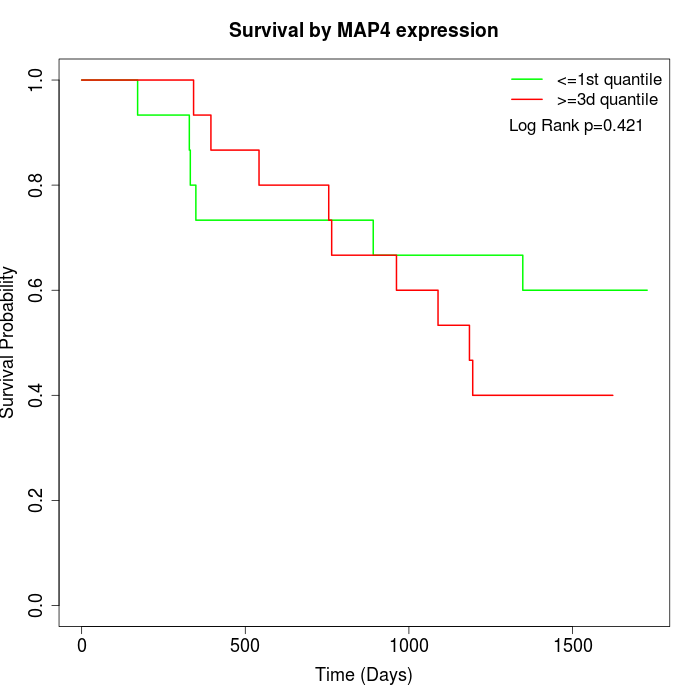
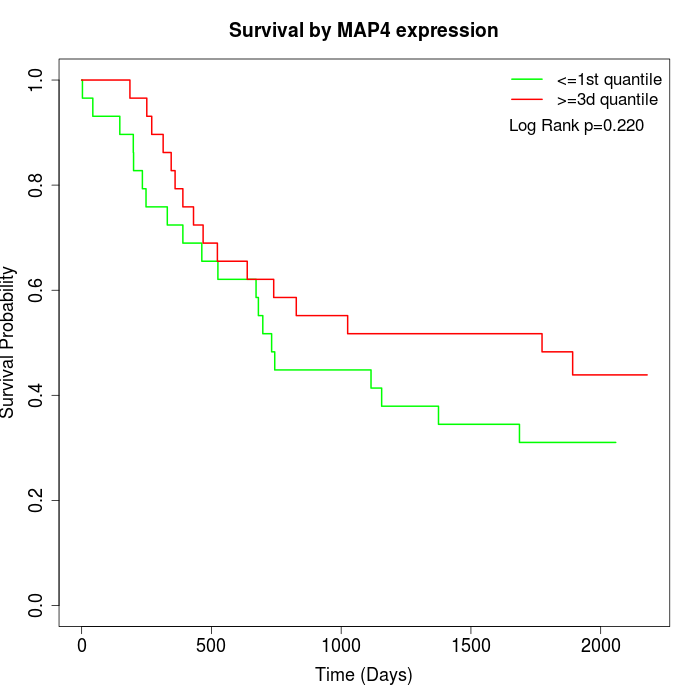
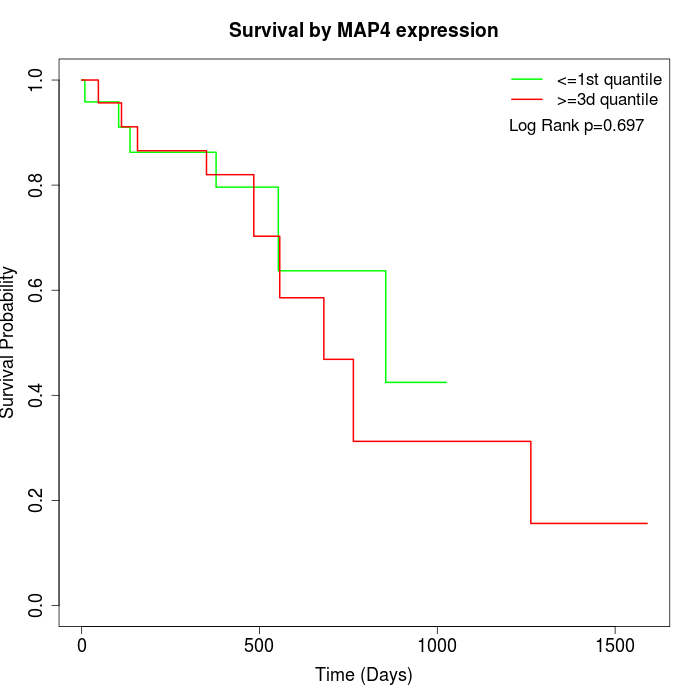
Survival by MAP4 expression:
Note: Click image to view full size file.
Copy number change of MAP4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP4 | 4134 | 0 | 18 | 12 | |
| GSE20123 | MAP4 | 4134 | 0 | 19 | 11 | |
| GSE43470 | MAP4 | 4134 | 0 | 19 | 24 | |
| GSE46452 | MAP4 | 4134 | 2 | 17 | 40 | |
| GSE47630 | MAP4 | 4134 | 2 | 23 | 15 | |
| GSE54993 | MAP4 | 4134 | 6 | 2 | 62 | |
| GSE54994 | MAP4 | 4134 | 0 | 36 | 17 | |
| GSE60625 | MAP4 | 4134 | 5 | 0 | 6 | |
| GSE74703 | MAP4 | 4134 | 0 | 15 | 21 | |
| GSE74704 | MAP4 | 4134 | 0 | 12 | 8 | |
| TCGA | MAP4 | 4134 | 0 | 77 | 19 |
Total number of gains: 15; Total number of losses: 238; Total Number of normals: 235.
Somatic mutations of MAP4:
Generating mutation plots.
Highly correlated genes for MAP4:
Showing top 20/604 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP4 | GLIS3 | 0.835733 | 3 | 0 | 3 |
| MAP4 | RGMB | 0.804246 | 3 | 0 | 3 |
| MAP4 | PRICKLE2 | 0.801177 | 3 | 0 | 3 |
| MAP4 | SH3RF3 | 0.799716 | 3 | 0 | 3 |
| MAP4 | C20orf194 | 0.779405 | 3 | 0 | 3 |
| MAP4 | HABP4 | 0.77096 | 6 | 0 | 6 |
| MAP4 | ARHGEF6 | 0.759694 | 4 | 0 | 4 |
| MAP4 | PDE12 | 0.755487 | 3 | 0 | 3 |
| MAP4 | LRRFIP2 | 0.755175 | 3 | 0 | 3 |
| MAP4 | SOCS2 | 0.754041 | 3 | 0 | 3 |
| MAP4 | DNALI1 | 0.75189 | 5 | 0 | 5 |
| MAP4 | KATNAL1 | 0.74646 | 3 | 0 | 3 |
| MAP4 | IGIP | 0.745271 | 3 | 0 | 3 |
| MAP4 | KRTAP19-2 | 0.744671 | 3 | 0 | 3 |
| MAP4 | EML1 | 0.742693 | 5 | 0 | 5 |
| MAP4 | ARMCX3 | 0.740536 | 3 | 0 | 3 |
| MAP4 | TGFB1I1 | 0.740131 | 7 | 0 | 7 |
| MAP4 | ITIH5 | 0.738419 | 3 | 0 | 3 |
| MAP4 | FYCO1 | 0.737964 | 6 | 0 | 6 |
| MAP4 | RAB11FIP2 | 0.736513 | 4 | 0 | 4 |
For details and further investigation, click here