| Full name: phosphodiesterase 12 | Alias Symbol: DKFZp667B1218|2'-PDE|3635 | ||
| Type: protein-coding gene | Cytoband: 3p14.3 | ||
| Entrez ID: 201626 | HGNC ID: HGNC:25386 | Ensembl Gene: ENSG00000174840 | OMIM ID: 616519 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PDE12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE12 | 201626 | 231839_at | -0.2203 | 0.5705 | |
| GSE20347 | PDE12 | 201626 | 214826_at | -0.0014 | 0.9879 | |
| GSE23400 | PDE12 | 201626 | 214826_at | -0.0999 | 0.0000 | |
| GSE26886 | PDE12 | 201626 | 231839_at | -0.5308 | 0.0011 | |
| GSE29001 | PDE12 | 201626 | 214826_at | -0.0689 | 0.7467 | |
| GSE38129 | PDE12 | 201626 | 214826_at | -0.1236 | 0.0148 | |
| GSE45670 | PDE12 | 201626 | 231839_at | -0.4223 | 0.0013 | |
| GSE53622 | PDE12 | 201626 | 59753 | -0.7513 | 0.0000 | |
| GSE53624 | PDE12 | 201626 | 61750 | -0.7911 | 0.0000 | |
| GSE63941 | PDE12 | 201626 | 231839_at | -0.6804 | 0.0728 | |
| GSE77861 | PDE12 | 201626 | 231839_at | -0.4610 | 0.0269 | |
| GSE97050 | PDE12 | 201626 | A_33_P3288017 | -0.1750 | 0.4004 | |
| SRP007169 | PDE12 | 201626 | RNAseq | -1.1647 | 0.0002 | |
| SRP008496 | PDE12 | 201626 | RNAseq | -0.7595 | 0.0002 | |
| SRP064894 | PDE12 | 201626 | RNAseq | -0.7719 | 0.0000 | |
| SRP133303 | PDE12 | 201626 | RNAseq | -0.3138 | 0.0069 | |
| SRP159526 | PDE12 | 201626 | RNAseq | -0.6550 | 0.0173 | |
| SRP193095 | PDE12 | 201626 | RNAseq | -0.4534 | 0.0000 | |
| SRP219564 | PDE12 | 201626 | RNAseq | -0.7166 | 0.0025 | |
| TCGA | PDE12 | 201626 | RNAseq | -0.2360 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
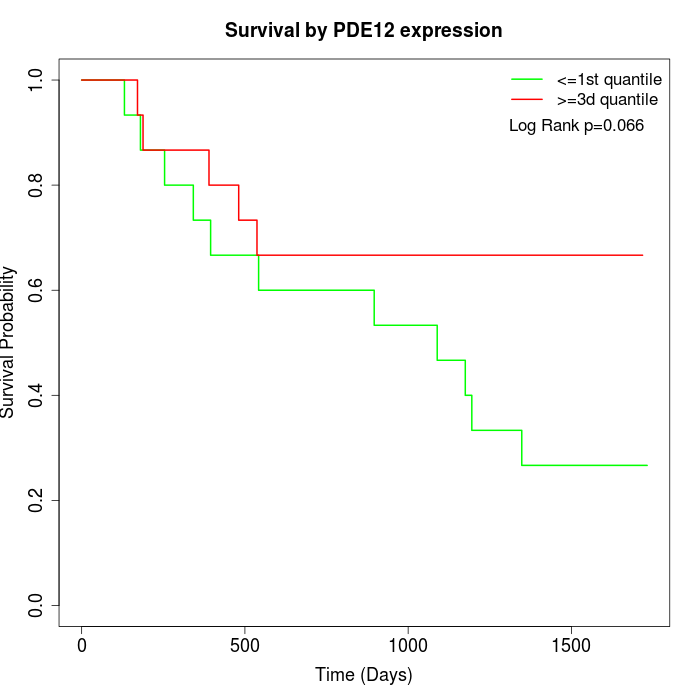
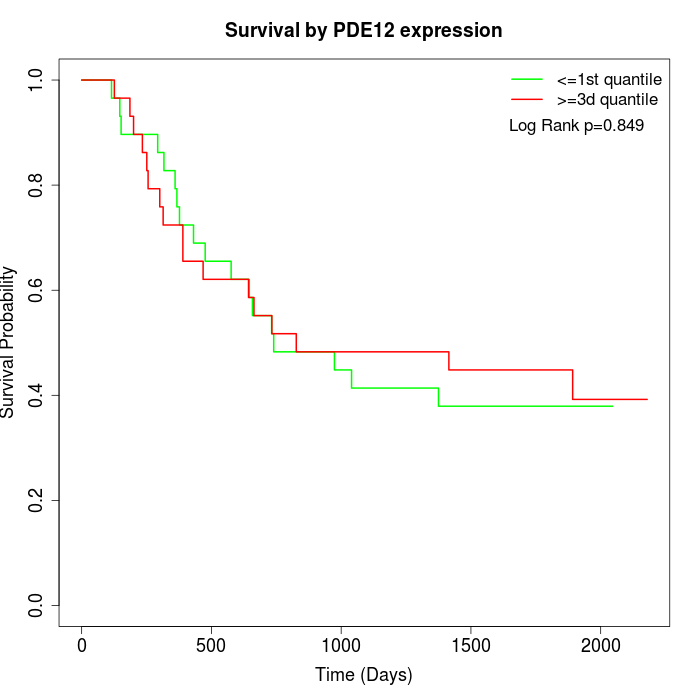
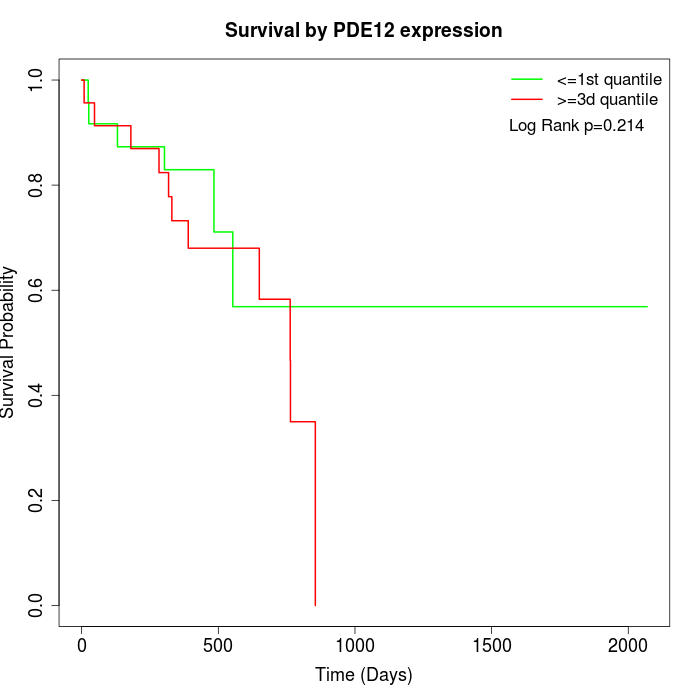
Survival by PDE12 expression:
Note: Click image to view full size file.
Copy number change of PDE12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE12 | 201626 | 0 | 17 | 13 | |
| GSE20123 | PDE12 | 201626 | 0 | 18 | 12 | |
| GSE43470 | PDE12 | 201626 | 0 | 17 | 26 | |
| GSE46452 | PDE12 | 201626 | 2 | 17 | 40 | |
| GSE47630 | PDE12 | 201626 | 1 | 24 | 15 | |
| GSE54993 | PDE12 | 201626 | 6 | 2 | 62 | |
| GSE54994 | PDE12 | 201626 | 0 | 32 | 21 | |
| GSE60625 | PDE12 | 201626 | 5 | 0 | 6 | |
| GSE74703 | PDE12 | 201626 | 0 | 14 | 22 | |
| GSE74704 | PDE12 | 201626 | 0 | 11 | 9 | |
| TCGA | PDE12 | 201626 | 0 | 79 | 17 |
Total number of gains: 14; Total number of losses: 231; Total Number of normals: 243.
Somatic mutations of PDE12:
Generating mutation plots.
Highly correlated genes for PDE12:
Showing top 20/1179 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE12 | CCNYL1 | 0.766042 | 5 | 0 | 5 |
| PDE12 | ESYT2 | 0.757028 | 4 | 0 | 4 |
| PDE12 | MAP4 | 0.755487 | 3 | 0 | 3 |
| PDE12 | SSBP3 | 0.75149 | 3 | 0 | 3 |
| PDE12 | HDHD2 | 0.74831 | 5 | 0 | 4 |
| PDE12 | FBXW5 | 0.744418 | 4 | 0 | 4 |
| PDE12 | ID4 | 0.736481 | 4 | 0 | 4 |
| PDE12 | PCDH11X | 0.732614 | 3 | 0 | 3 |
| PDE12 | BTAF1 | 0.730153 | 4 | 0 | 4 |
| PDE12 | DCAF10 | 0.724178 | 3 | 0 | 3 |
| PDE12 | OGFOD1 | 0.722266 | 3 | 0 | 3 |
| PDE12 | BBIP1 | 0.722076 | 5 | 0 | 5 |
| PDE12 | RUFY1 | 0.714903 | 6 | 0 | 6 |
| PDE12 | CCDC12 | 0.714299 | 6 | 0 | 6 |
| PDE12 | TRAPPC6B | 0.708754 | 3 | 0 | 3 |
| PDE12 | SUGT1 | 0.708358 | 4 | 0 | 3 |
| PDE12 | SLC19A2 | 0.707628 | 3 | 0 | 3 |
| PDE12 | SOCS6 | 0.706147 | 6 | 0 | 5 |
| PDE12 | SLAMF7 | 0.703345 | 3 | 0 | 3 |
| PDE12 | RBM20 | 0.702653 | 4 | 0 | 4 |
For details and further investigation, click here