| Full name: IgA inducing protein | Alias Symbol: LOC492311 | ||
| Type: protein-coding gene | Cytoband: 5q31.3 | ||
| Entrez ID: 492311 | HGNC ID: HGNC:33847 | Ensembl Gene: ENSG00000182700 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of IGIP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IGIP | 492311 | 226977_at | -0.4600 | 0.5119 | |
| GSE26886 | IGIP | 492311 | 226977_at | -0.5321 | 0.0454 | |
| GSE45670 | IGIP | 492311 | 226977_at | -0.4550 | 0.0166 | |
| GSE53622 | IGIP | 492311 | 18502 | -0.9230 | 0.0000 | |
| GSE53624 | IGIP | 492311 | 18502 | -0.8103 | 0.0000 | |
| GSE63941 | IGIP | 492311 | 226977_at | -1.6568 | 0.0036 | |
| GSE77861 | IGIP | 492311 | 226977_at | -0.1910 | 0.4152 | |
| GSE97050 | IGIP | 492311 | A_24_P110558 | -0.7322 | 0.1476 | |
| SRP007169 | IGIP | 492311 | RNAseq | 0.2062 | 0.6538 | |
| SRP008496 | IGIP | 492311 | RNAseq | 0.2748 | 0.4063 | |
| SRP064894 | IGIP | 492311 | RNAseq | 0.0951 | 0.5722 | |
| SRP133303 | IGIP | 492311 | RNAseq | -0.1251 | 0.6532 | |
| SRP159526 | IGIP | 492311 | RNAseq | -0.5467 | 0.0278 | |
| SRP193095 | IGIP | 492311 | RNAseq | -0.6507 | 0.0000 | |
| SRP219564 | IGIP | 492311 | RNAseq | -0.1444 | 0.7537 |
Upregulated datasets: 0; Downregulated datasets: 1.
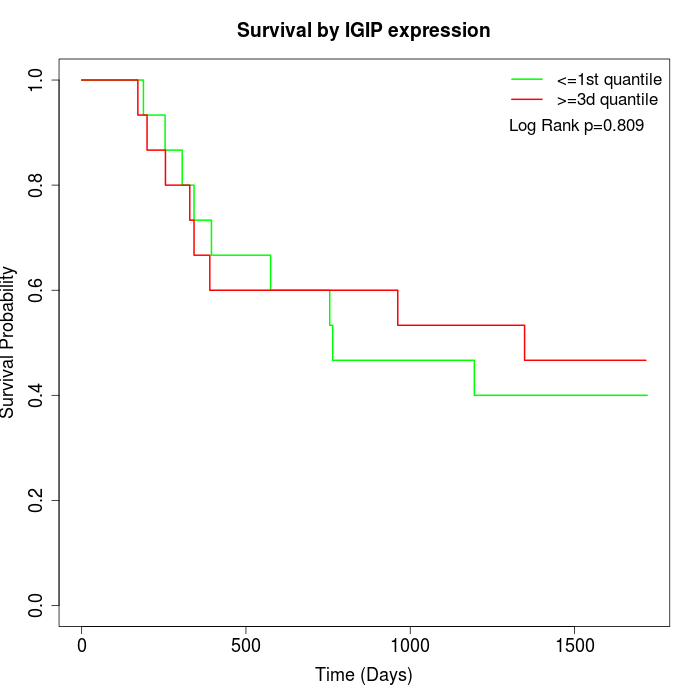
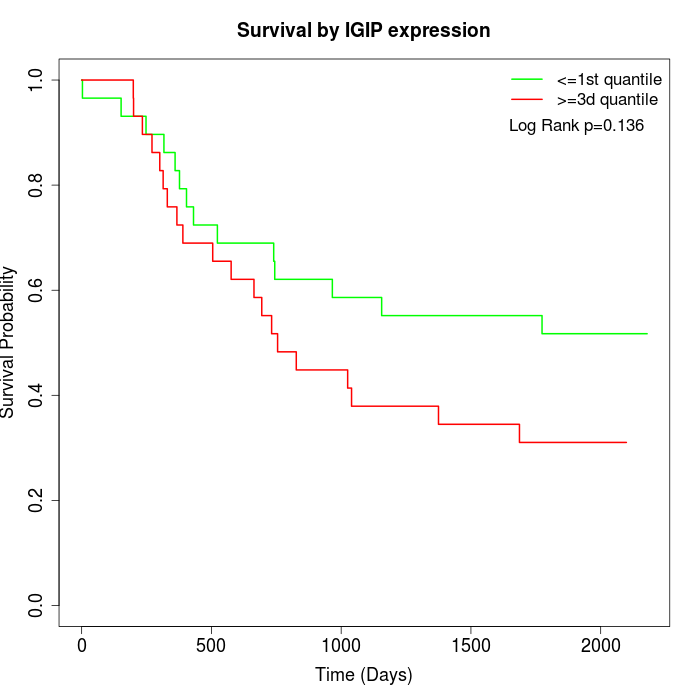
Survival by IGIP expression:
Note: Click image to view full size file.
Copy number change of IGIP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IGIP | 492311 | 3 | 11 | 16 | |
| GSE20123 | IGIP | 492311 | 4 | 11 | 15 | |
| GSE43470 | IGIP | 492311 | 4 | 8 | 31 | |
| GSE46452 | IGIP | 492311 | 0 | 27 | 32 | |
| GSE47630 | IGIP | 492311 | 0 | 20 | 20 | |
| GSE54993 | IGIP | 492311 | 9 | 1 | 60 | |
| GSE54994 | IGIP | 492311 | 2 | 14 | 37 | |
| GSE60625 | IGIP | 492311 | 0 | 0 | 11 | |
| GSE74703 | IGIP | 492311 | 3 | 6 | 27 | |
| GSE74704 | IGIP | 492311 | 3 | 5 | 12 | |
| TCGA | IGIP | 492311 | 4 | 37 | 55 |
Total number of gains: 32; Total number of losses: 140; Total Number of normals: 316.
Somatic mutations of IGIP:
Generating mutation plots.
Highly correlated genes for IGIP:
Showing top 20/1234 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IGIP | ZNF225 | 0.816925 | 3 | 0 | 3 |
| IGIP | C5orf24 | 0.816861 | 6 | 0 | 6 |
| IGIP | PURA | 0.808615 | 8 | 0 | 8 |
| IGIP | RAB23 | 0.78937 | 3 | 0 | 3 |
| IGIP | TNFSF12 | 0.787929 | 3 | 0 | 3 |
| IGIP | WDR48 | 0.777955 | 3 | 0 | 3 |
| IGIP | FLNA | 0.777319 | 3 | 0 | 3 |
| IGIP | NPY1R | 0.773301 | 3 | 0 | 3 |
| IGIP | C1QTNF7 | 0.770719 | 5 | 0 | 5 |
| IGIP | KIF5C | 0.768691 | 3 | 0 | 3 |
| IGIP | PDS5B | 0.76223 | 3 | 0 | 3 |
| IGIP | FZD7 | 0.758149 | 3 | 0 | 3 |
| IGIP | TRPC6 | 0.757935 | 3 | 0 | 3 |
| IGIP | DENND5A | 0.752981 | 3 | 0 | 3 |
| IGIP | PSIP1 | 0.752549 | 3 | 0 | 3 |
| IGIP | CDS2 | 0.751951 | 3 | 0 | 3 |
| IGIP | NFASC | 0.75013 | 4 | 0 | 4 |
| IGIP | GPRASP2 | 0.749606 | 3 | 0 | 3 |
| IGIP | CNST | 0.747639 | 3 | 0 | 3 |
| IGIP | MAP4 | 0.745271 | 3 | 0 | 3 |
For details and further investigation, click here