| Full name: MAP7 domain containing 3 | Alias Symbol: FLJ12649 | ||
| Type: protein-coding gene | Cytoband: Xq26.3 | ||
| Entrez ID: 79649 | HGNC ID: HGNC:25742 | Ensembl Gene: ENSG00000129680 | OMIM ID: 300930 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP7D3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP7D3 | 79649 | 219626_at | 0.0243 | 0.9563 | |
| GSE20347 | MAP7D3 | 79649 | 219576_at | 0.2855 | 0.0014 | |
| GSE23400 | MAP7D3 | 79649 | 219626_at | 0.0866 | 0.0478 | |
| GSE26886 | MAP7D3 | 79649 | 219626_at | 0.0616 | 0.7542 | |
| GSE29001 | MAP7D3 | 79649 | 219626_at | 0.4646 | 0.0224 | |
| GSE38129 | MAP7D3 | 79649 | 219576_at | 0.0918 | 0.6435 | |
| GSE45670 | MAP7D3 | 79649 | 219626_at | -0.1221 | 0.5391 | |
| GSE53622 | MAP7D3 | 79649 | 144260 | 0.0893 | 0.4192 | |
| GSE53624 | MAP7D3 | 79649 | 144260 | 0.2905 | 0.0083 | |
| GSE63941 | MAP7D3 | 79649 | 219626_at | -0.8554 | 0.1200 | |
| GSE77861 | MAP7D3 | 79649 | 219626_at | 0.1925 | 0.0610 | |
| GSE97050 | MAP7D3 | 79649 | A_23_P11160 | -0.1662 | 0.6390 | |
| SRP007169 | MAP7D3 | 79649 | RNAseq | 1.9598 | 0.0021 | |
| SRP008496 | MAP7D3 | 79649 | RNAseq | 2.6540 | 0.0000 | |
| SRP064894 | MAP7D3 | 79649 | RNAseq | 0.5850 | 0.0031 | |
| SRP133303 | MAP7D3 | 79649 | RNAseq | 0.3751 | 0.0502 | |
| SRP159526 | MAP7D3 | 79649 | RNAseq | 0.1046 | 0.7433 | |
| SRP193095 | MAP7D3 | 79649 | RNAseq | 0.9032 | 0.0000 | |
| SRP219564 | MAP7D3 | 79649 | RNAseq | -0.0349 | 0.9411 | |
| TCGA | MAP7D3 | 79649 | RNAseq | 0.1300 | 0.1391 |
Upregulated datasets: 2; Downregulated datasets: 0.
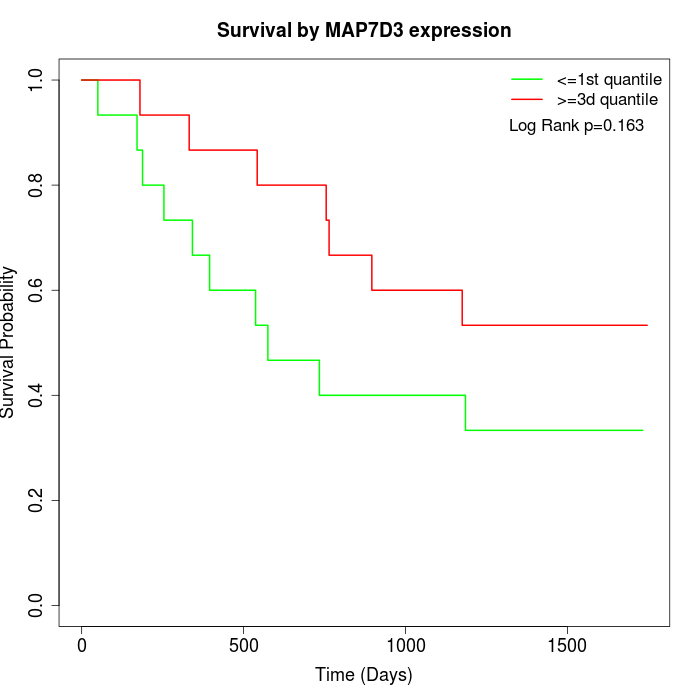
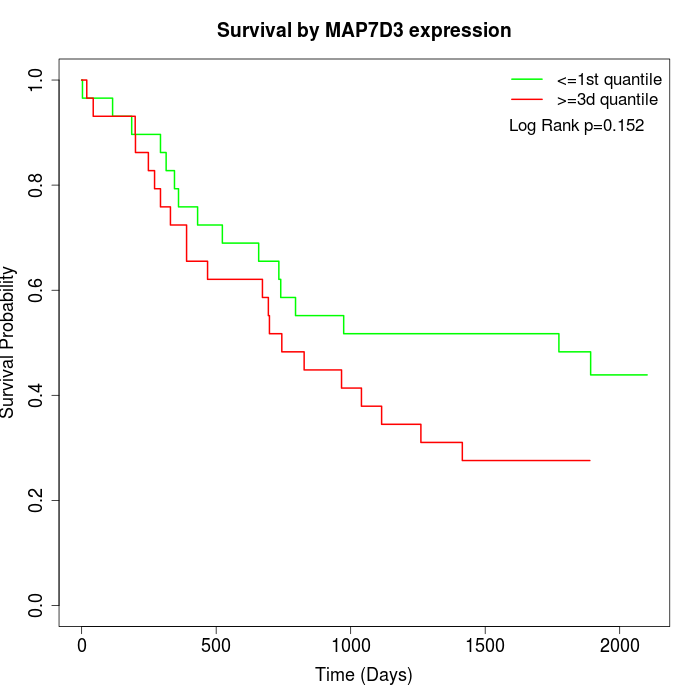
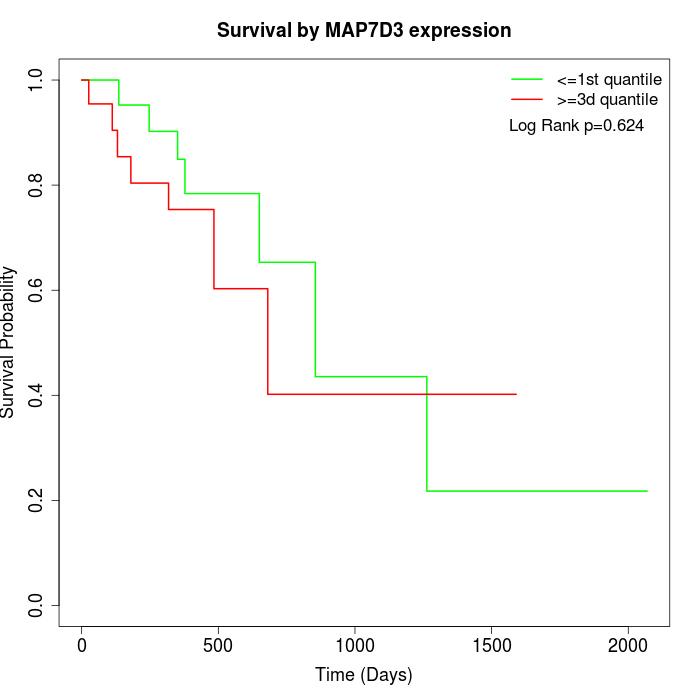
Survival by MAP7D3 expression:
Note: Click image to view full size file.
Copy number change of MAP7D3:
No record found for this gene.
Somatic mutations of MAP7D3:
Generating mutation plots.
Highly correlated genes for MAP7D3:
Showing top 20/337 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP7D3 | CACNA1H | 0.706758 | 3 | 0 | 3 |
| MAP7D3 | ZNF275 | 0.68703 | 4 | 0 | 4 |
| MAP7D3 | MMGT1 | 0.686939 | 5 | 0 | 5 |
| MAP7D3 | FUNDC2 | 0.677217 | 4 | 0 | 3 |
| MAP7D3 | INPP4A | 0.649889 | 4 | 0 | 3 |
| MAP7D3 | SGCE | 0.646 | 6 | 0 | 5 |
| MAP7D3 | DNAJC8 | 0.643861 | 5 | 0 | 5 |
| MAP7D3 | PKIG | 0.642828 | 6 | 0 | 5 |
| MAP7D3 | KLHL42 | 0.641522 | 5 | 0 | 3 |
| MAP7D3 | LENG1 | 0.64058 | 3 | 0 | 3 |
| MAP7D3 | FGL2 | 0.637917 | 3 | 0 | 3 |
| MAP7D3 | DCHS1 | 0.636943 | 5 | 0 | 4 |
| MAP7D3 | KCNJ8 | 0.636314 | 7 | 0 | 5 |
| MAP7D3 | DAPK1 | 0.634548 | 3 | 0 | 3 |
| MAP7D3 | MCAM | 0.632436 | 8 | 0 | 5 |
| MAP7D3 | MAP9 | 0.626653 | 5 | 0 | 3 |
| MAP7D3 | C1orf216 | 0.626452 | 10 | 0 | 9 |
| MAP7D3 | COPRS | 0.625276 | 6 | 0 | 5 |
| MAP7D3 | FAM210B | 0.620252 | 3 | 0 | 3 |
| MAP7D3 | LRRC49 | 0.619103 | 9 | 0 | 9 |
For details and further investigation, click here