| Full name: microtubule associated protein 9 | Alias Symbol: ASAP|FLJ21159 | ||
| Type: protein-coding gene | Cytoband: 4q32.1 | ||
| Entrez ID: 79884 | HGNC ID: HGNC:26118 | Ensembl Gene: ENSG00000164114 | OMIM ID: 610070 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP9 | 79884 | 228423_at | -0.1148 | 0.9574 | |
| GSE20347 | MAP9 | 79884 | 220145_at | 0.0286 | 0.8812 | |
| GSE23400 | MAP9 | 79884 | 220145_at | 0.0212 | 0.3183 | |
| GSE26886 | MAP9 | 79884 | 228423_at | 1.9967 | 0.0020 | |
| GSE29001 | MAP9 | 79884 | 220145_at | 0.0670 | 0.8484 | |
| GSE38129 | MAP9 | 79884 | 220145_at | -0.2907 | 0.5213 | |
| GSE45670 | MAP9 | 79884 | 228423_at | -0.8948 | 0.0777 | |
| GSE53622 | MAP9 | 79884 | 80048 | 0.2193 | 0.2174 | |
| GSE53624 | MAP9 | 79884 | 80048 | 0.2019 | 0.1731 | |
| GSE63941 | MAP9 | 79884 | 228423_at | -3.5301 | 0.0122 | |
| GSE77861 | MAP9 | 79884 | 239415_at | 0.1139 | 0.4510 | |
| GSE97050 | MAP9 | 79884 | A_32_P50066 | -0.5141 | 0.3138 | |
| SRP007169 | MAP9 | 79884 | RNAseq | 0.7209 | 0.2132 | |
| SRP008496 | MAP9 | 79884 | RNAseq | 2.0655 | 0.0007 | |
| SRP064894 | MAP9 | 79884 | RNAseq | 0.6525 | 0.0562 | |
| SRP133303 | MAP9 | 79884 | RNAseq | 0.6331 | 0.1228 | |
| SRP159526 | MAP9 | 79884 | RNAseq | 1.4806 | 0.0039 | |
| SRP193095 | MAP9 | 79884 | RNAseq | 0.6420 | 0.0395 | |
| SRP219564 | MAP9 | 79884 | RNAseq | -0.0362 | 0.9656 | |
| TCGA | MAP9 | 79884 | RNAseq | -0.4579 | 0.0182 |
Upregulated datasets: 3; Downregulated datasets: 1.
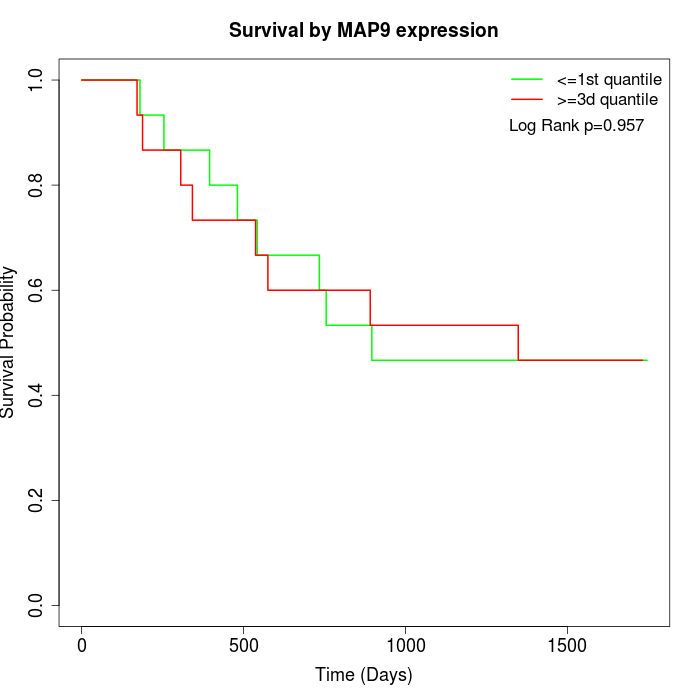
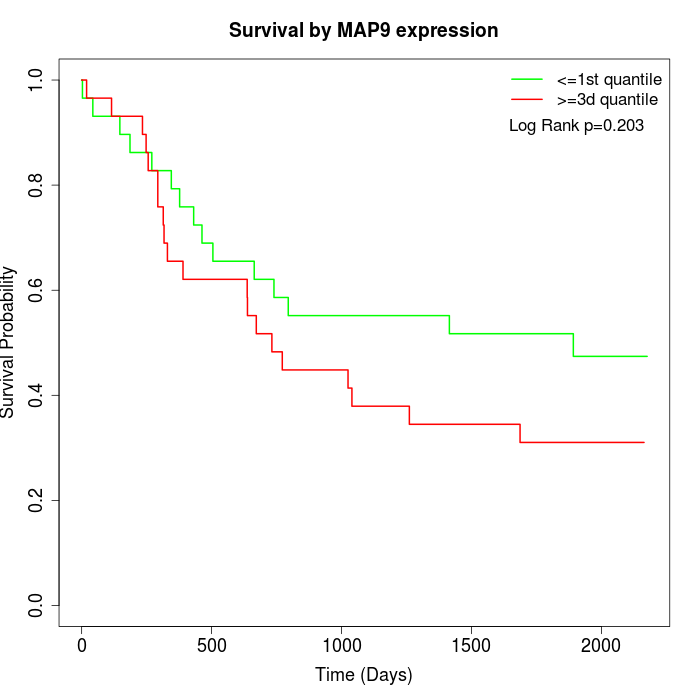
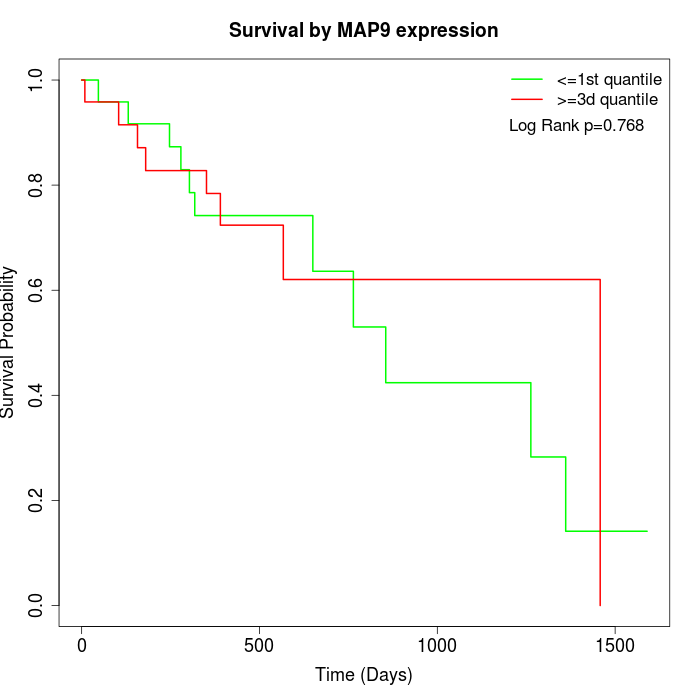
Survival by MAP9 expression:
Note: Click image to view full size file.
Copy number change of MAP9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP9 | 79884 | 0 | 14 | 16 | |
| GSE20123 | MAP9 | 79884 | 0 | 14 | 16 | |
| GSE43470 | MAP9 | 79884 | 0 | 14 | 29 | |
| GSE46452 | MAP9 | 79884 | 1 | 36 | 22 | |
| GSE47630 | MAP9 | 79884 | 0 | 23 | 17 | |
| GSE54993 | MAP9 | 79884 | 10 | 0 | 60 | |
| GSE54994 | MAP9 | 79884 | 1 | 12 | 40 | |
| GSE60625 | MAP9 | 79884 | 0 | 1 | 10 | |
| GSE74703 | MAP9 | 79884 | 0 | 12 | 24 | |
| GSE74704 | MAP9 | 79884 | 0 | 7 | 13 | |
| TCGA | MAP9 | 79884 | 10 | 35 | 51 |
Total number of gains: 22; Total number of losses: 168; Total Number of normals: 298.
Somatic mutations of MAP9:
Generating mutation plots.
Highly correlated genes for MAP9:
Showing top 20/897 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP9 | USP9X | 0.757867 | 3 | 0 | 3 |
| MAP9 | DDHD1 | 0.7424 | 3 | 0 | 3 |
| MAP9 | ZCCHC7 | 0.724213 | 3 | 0 | 3 |
| MAP9 | GNB5 | 0.724074 | 3 | 0 | 3 |
| MAP9 | ABCB7 | 0.721437 | 3 | 0 | 3 |
| MAP9 | DNAJC8 | 0.720676 | 4 | 0 | 3 |
| MAP9 | CKMT2 | 0.720322 | 3 | 0 | 3 |
| MAP9 | BDNF | 0.715536 | 3 | 0 | 3 |
| MAP9 | MYADM | 0.712156 | 4 | 0 | 4 |
| MAP9 | NME5 | 0.7113 | 5 | 0 | 5 |
| MAP9 | TET1 | 0.710524 | 3 | 0 | 3 |
| MAP9 | SELENBP1 | 0.708755 | 3 | 0 | 3 |
| MAP9 | CENPV | 0.704457 | 5 | 0 | 4 |
| MAP9 | FLNA | 0.700265 | 5 | 0 | 5 |
| MAP9 | NHS | 0.700133 | 4 | 0 | 3 |
| MAP9 | SGCE | 0.699767 | 9 | 0 | 8 |
| MAP9 | UBXN1 | 0.6991 | 3 | 0 | 3 |
| MAP9 | RCAN2 | 0.698388 | 5 | 0 | 5 |
| MAP9 | ARHGAP6 | 0.697829 | 5 | 0 | 4 |
| MAP9 | FXYD1 | 0.693472 | 3 | 0 | 3 |
For details and further investigation, click here