| Full name: microtubule affinity regulating kinase 1 | Alias Symbol: MARK|PAR-1C | ||
| Type: protein-coding gene | Cytoband: 1q41 | ||
| Entrez ID: 4139 | HGNC ID: HGNC:6896 | Ensembl Gene: ENSG00000116141 | OMIM ID: 606511 |
| Drug and gene relationship at DGIdb | |||
Expression of MARK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MARK1 | 4139 | 221047_s_at | 0.7623 | 0.1935 | |
| GSE20347 | MARK1 | 4139 | 221047_s_at | 0.6404 | 0.0005 | |
| GSE23400 | MARK1 | 4139 | 221047_s_at | 0.4742 | 0.0000 | |
| GSE26886 | MARK1 | 4139 | 221047_s_at | 1.8365 | 0.0000 | |
| GSE29001 | MARK1 | 4139 | 221047_s_at | 0.8614 | 0.0096 | |
| GSE38129 | MARK1 | 4139 | 221047_s_at | 0.8040 | 0.0000 | |
| GSE45670 | MARK1 | 4139 | 221047_s_at | 0.7376 | 0.0006 | |
| GSE53622 | MARK1 | 4139 | 74745 | 0.2764 | 0.0580 | |
| GSE53624 | MARK1 | 4139 | 74745 | 0.3444 | 0.0085 | |
| GSE63941 | MARK1 | 4139 | 221047_s_at | 2.6226 | 0.0048 | |
| GSE77861 | MARK1 | 4139 | 221047_s_at | 0.9680 | 0.0015 | |
| GSE97050 | MARK1 | 4139 | A_24_P179585 | 0.1260 | 0.5725 | |
| SRP007169 | MARK1 | 4139 | RNAseq | 1.0721 | 0.0616 | |
| SRP008496 | MARK1 | 4139 | RNAseq | 1.3419 | 0.0056 | |
| SRP064894 | MARK1 | 4139 | RNAseq | 0.3820 | 0.0828 | |
| SRP133303 | MARK1 | 4139 | RNAseq | 0.9925 | 0.0000 | |
| SRP159526 | MARK1 | 4139 | RNAseq | 0.9485 | 0.0029 | |
| SRP193095 | MARK1 | 4139 | RNAseq | 0.9498 | 0.0010 | |
| SRP219564 | MARK1 | 4139 | RNAseq | 0.3714 | 0.2031 | |
| TCGA | MARK1 | 4139 | RNAseq | 0.3211 | 0.0010 |
Upregulated datasets: 3; Downregulated datasets: 0.
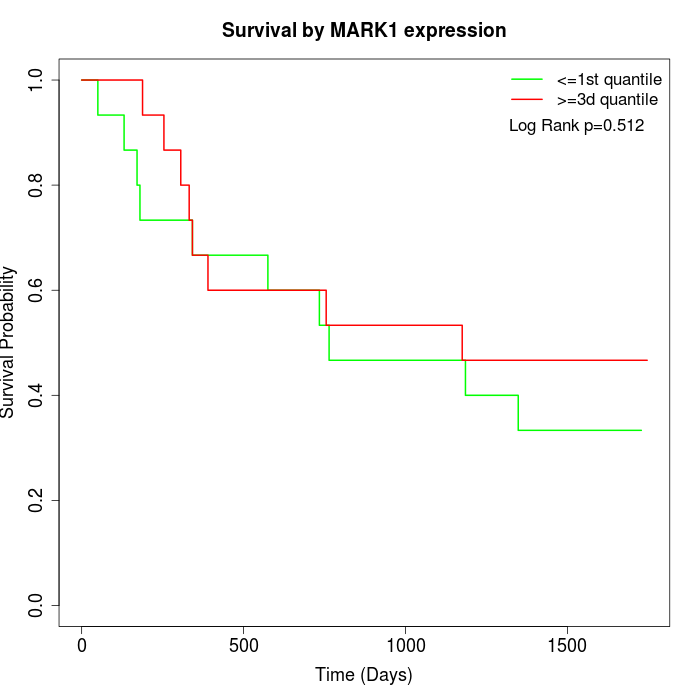
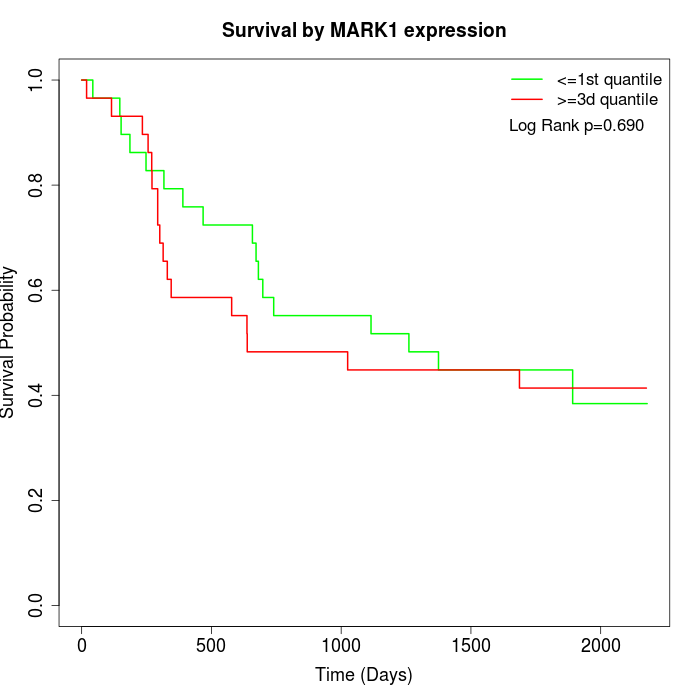
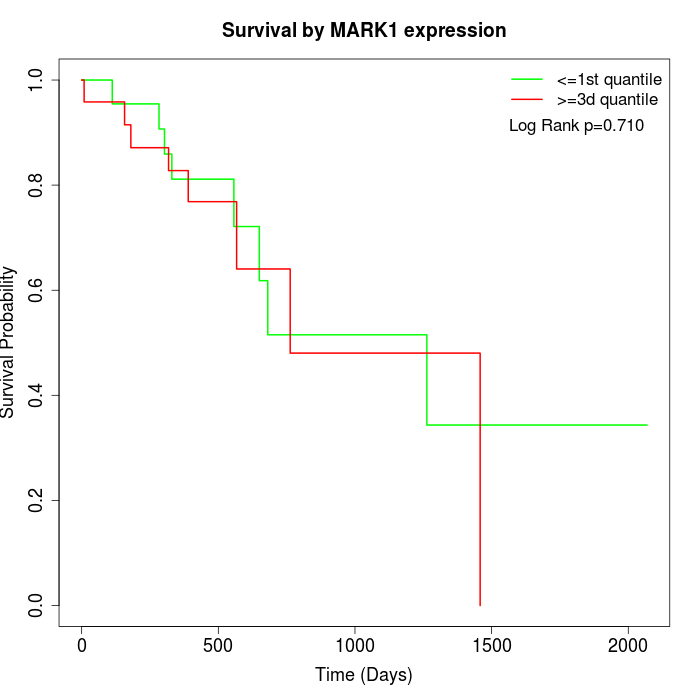
Survival by MARK1 expression:
Note: Click image to view full size file.
Copy number change of MARK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MARK1 | 4139 | 10 | 0 | 20 | |
| GSE20123 | MARK1 | 4139 | 10 | 0 | 20 | |
| GSE43470 | MARK1 | 4139 | 8 | 1 | 34 | |
| GSE46452 | MARK1 | 4139 | 3 | 1 | 55 | |
| GSE47630 | MARK1 | 4139 | 15 | 0 | 25 | |
| GSE54993 | MARK1 | 4139 | 0 | 6 | 64 | |
| GSE54994 | MARK1 | 4139 | 15 | 0 | 38 | |
| GSE60625 | MARK1 | 4139 | 0 | 0 | 11 | |
| GSE74703 | MARK1 | 4139 | 8 | 1 | 27 | |
| GSE74704 | MARK1 | 4139 | 4 | 0 | 16 | |
| TCGA | MARK1 | 4139 | 45 | 3 | 48 |
Total number of gains: 118; Total number of losses: 12; Total Number of normals: 358.
Somatic mutations of MARK1:
Generating mutation plots.
Highly correlated genes for MARK1:
Showing top 20/1472 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MARK1 | C15orf61 | 0.830443 | 3 | 0 | 3 |
| MARK1 | RILPL2 | 0.819457 | 3 | 0 | 3 |
| MARK1 | NTPCR | 0.806449 | 5 | 0 | 5 |
| MARK1 | FAM89B | 0.800931 | 3 | 0 | 3 |
| MARK1 | MRPS26 | 0.800317 | 3 | 0 | 3 |
| MARK1 | WDR66 | 0.773571 | 5 | 0 | 5 |
| MARK1 | RPS27A | 0.760703 | 3 | 0 | 3 |
| MARK1 | WDR81 | 0.757589 | 4 | 0 | 4 |
| MARK1 | TMEM138 | 0.757138 | 5 | 0 | 5 |
| MARK1 | EID2 | 0.756765 | 3 | 0 | 3 |
| MARK1 | PIGH | 0.753973 | 3 | 0 | 3 |
| MARK1 | RILPL1 | 0.751952 | 3 | 0 | 3 |
| MARK1 | GMCL1 | 0.745025 | 3 | 0 | 3 |
| MARK1 | MRPS31 | 0.741655 | 3 | 0 | 3 |
| MARK1 | WDR72 | 0.740972 | 4 | 0 | 4 |
| MARK1 | EIF3M | 0.737121 | 3 | 0 | 3 |
| MARK1 | ANKRD11 | 0.736636 | 3 | 0 | 3 |
| MARK1 | NGF | 0.735248 | 3 | 0 | 3 |
| MARK1 | WDR4 | 0.734231 | 3 | 0 | 3 |
| MARK1 | MSTO1 | 0.732778 | 5 | 0 | 4 |
For details and further investigation, click here