| Full name: kelch like family member 24 | Alias Symbol: DRE1|FLJ20059 | ||
| Type: protein-coding gene | Cytoband: 3q27.1 | ||
| Entrez ID: 54800 | HGNC ID: HGNC:25947 | Ensembl Gene: ENSG00000114796 | OMIM ID: 611295 |
| Drug and gene relationship at DGIdb | |||
Expression of KLHL24:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KLHL24 | 54800 | 226158_at | 0.3521 | 0.6041 | |
| GSE20347 | KLHL24 | 54800 | 206551_x_at | 0.5066 | 0.0066 | |
| GSE23400 | KLHL24 | 54800 | 206551_x_at | 0.1411 | 0.0103 | |
| GSE26886 | KLHL24 | 54800 | 206551_x_at | 0.4528 | 0.0347 | |
| GSE29001 | KLHL24 | 54800 | 206551_x_at | 0.2973 | 0.0370 | |
| GSE38129 | KLHL24 | 54800 | 206551_x_at | 0.2372 | 0.1074 | |
| GSE45670 | KLHL24 | 54800 | 206551_x_at | 0.2653 | 0.0551 | |
| GSE53622 | KLHL24 | 54800 | 126543 | 0.5131 | 0.0004 | |
| GSE53624 | KLHL24 | 54800 | 126543 | 0.5514 | 0.0000 | |
| GSE63941 | KLHL24 | 54800 | 221985_at | 0.4790 | 0.5830 | |
| GSE77861 | KLHL24 | 54800 | 206551_x_at | 0.4724 | 0.1076 | |
| GSE97050 | KLHL24 | 54800 | A_24_P521994 | -0.2137 | 0.4039 | |
| SRP007169 | KLHL24 | 54800 | RNAseq | 1.1792 | 0.0354 | |
| SRP008496 | KLHL24 | 54800 | RNAseq | 1.1993 | 0.0002 | |
| SRP064894 | KLHL24 | 54800 | RNAseq | 0.2544 | 0.3409 | |
| SRP133303 | KLHL24 | 54800 | RNAseq | 0.7175 | 0.0000 | |
| SRP159526 | KLHL24 | 54800 | RNAseq | 1.5295 | 0.0000 | |
| SRP193095 | KLHL24 | 54800 | RNAseq | 0.5109 | 0.0069 | |
| SRP219564 | KLHL24 | 54800 | RNAseq | 0.1352 | 0.6155 | |
| TCGA | KLHL24 | 54800 | RNAseq | 0.0337 | 0.6522 |
Upregulated datasets: 3; Downregulated datasets: 0.
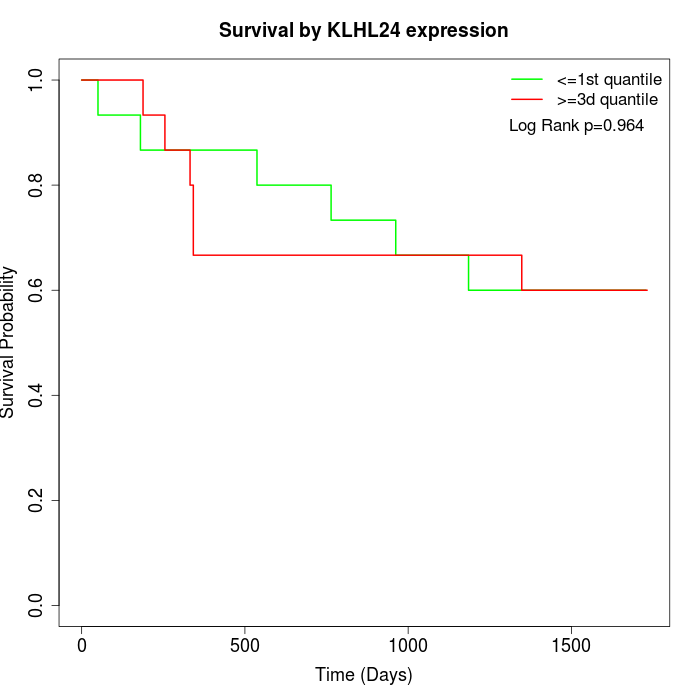
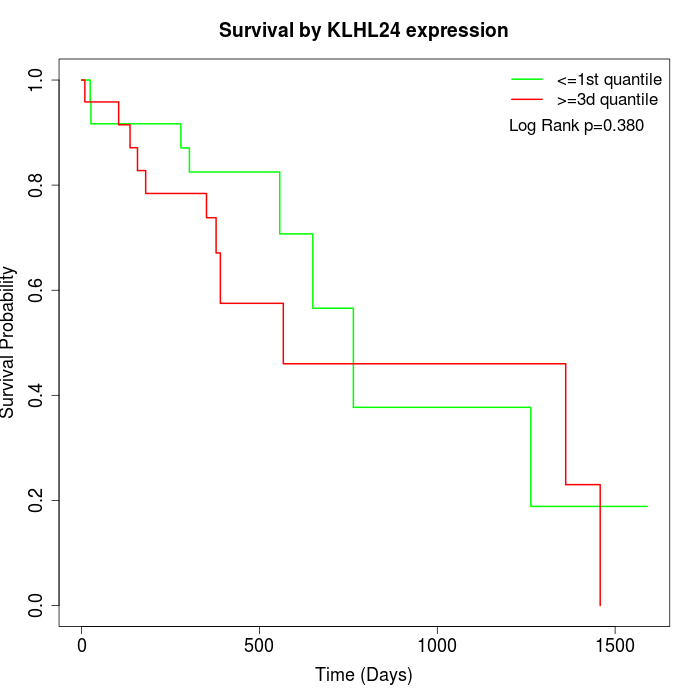
Survival by KLHL24 expression:
Note: Click image to view full size file.
Copy number change of KLHL24:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KLHL24 | 54800 | 25 | 0 | 5 | |
| GSE20123 | KLHL24 | 54800 | 25 | 0 | 5 | |
| GSE43470 | KLHL24 | 54800 | 24 | 0 | 19 | |
| GSE46452 | KLHL24 | 54800 | 19 | 1 | 39 | |
| GSE47630 | KLHL24 | 54800 | 27 | 2 | 11 | |
| GSE54993 | KLHL24 | 54800 | 1 | 19 | 50 | |
| GSE54994 | KLHL24 | 54800 | 40 | 0 | 13 | |
| GSE60625 | KLHL24 | 54800 | 0 | 6 | 5 | |
| GSE74703 | KLHL24 | 54800 | 20 | 0 | 16 | |
| GSE74704 | KLHL24 | 54800 | 17 | 0 | 3 | |
| TCGA | KLHL24 | 54800 | 77 | 0 | 19 |
Total number of gains: 275; Total number of losses: 28; Total Number of normals: 185.
Somatic mutations of KLHL24:
Generating mutation plots.
Highly correlated genes for KLHL24:
Showing top 20/478 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KLHL24 | FGFR1 | 0.841672 | 5 | 0 | 5 |
| KLHL24 | MAGI1 | 0.837324 | 4 | 0 | 4 |
| KLHL24 | ZC3H7B | 0.819019 | 8 | 0 | 7 |
| KLHL24 | USP34 | 0.791641 | 7 | 0 | 6 |
| KLHL24 | GTSE1 | 0.782844 | 7 | 0 | 7 |
| KLHL24 | FIP1L1 | 0.77532 | 3 | 0 | 3 |
| KLHL24 | TAOK1 | 0.770711 | 6 | 0 | 5 |
| KLHL24 | SH2B2 | 0.760346 | 8 | 0 | 8 |
| KLHL24 | ALMS1 | 0.758859 | 11 | 0 | 9 |
| KLHL24 | CCDC59 | 0.74833 | 3 | 0 | 3 |
| KLHL24 | CTCF | 0.74509 | 3 | 0 | 3 |
| KLHL24 | TYW1 | 0.736984 | 3 | 0 | 3 |
| KLHL24 | TBRG4 | 0.734738 | 3 | 0 | 3 |
| KLHL24 | PWWP2A | 0.733423 | 3 | 0 | 3 |
| KLHL24 | TRMT6 | 0.729905 | 3 | 0 | 3 |
| KLHL24 | POLR1B | 0.72229 | 8 | 0 | 6 |
| KLHL24 | MTRF1L | 0.717769 | 6 | 0 | 5 |
| KLHL24 | PMPCB | 0.716593 | 4 | 0 | 3 |
| KLHL24 | ANKS3 | 0.713307 | 3 | 0 | 3 |
| KLHL24 | METTL23 | 0.71283 | 3 | 0 | 3 |
For details and further investigation, click here