| Full name: muscleblind like splicing regulator 3 | Alias Symbol: CHCR|FLJ11316|MBLX39|MBXL | ||
| Type: protein-coding gene | Cytoband: Xq26.2 | ||
| Entrez ID: 55796 | HGNC ID: HGNC:20564 | Ensembl Gene: ENSG00000076770 | OMIM ID: 300413 |
| Drug and gene relationship at DGIdb | |||
Expression of MBNL3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MBNL3 | 55796 | 229498_at | -0.3707 | 0.5862 | |
| GSE20347 | MBNL3 | 55796 | 219814_at | -0.2750 | 0.0019 | |
| GSE23400 | MBNL3 | 55796 | 219814_at | -0.0798 | 0.0065 | |
| GSE26886 | MBNL3 | 55796 | 229498_at | -0.8608 | 0.0002 | |
| GSE29001 | MBNL3 | 55796 | 219814_at | -0.4176 | 0.1440 | |
| GSE38129 | MBNL3 | 55796 | 219814_at | -0.1640 | 0.1454 | |
| GSE45670 | MBNL3 | 55796 | 229498_at | -0.6324 | 0.0390 | |
| GSE53622 | MBNL3 | 55796 | 12262 | -0.1763 | 0.0001 | |
| GSE53624 | MBNL3 | 55796 | 12262 | -0.1441 | 0.0006 | |
| GSE63941 | MBNL3 | 55796 | 229498_at | -0.5988 | 0.6535 | |
| GSE77861 | MBNL3 | 55796 | 229498_at | -1.4631 | 0.0018 | |
| GSE97050 | MBNL3 | 55796 | A_33_P3414202 | 0.0919 | 0.6996 | |
| SRP007169 | MBNL3 | 55796 | RNAseq | -1.2446 | 0.0155 | |
| SRP008496 | MBNL3 | 55796 | RNAseq | -1.0983 | 0.0002 | |
| SRP064894 | MBNL3 | 55796 | RNAseq | -1.0306 | 0.0000 | |
| SRP133303 | MBNL3 | 55796 | RNAseq | -0.6753 | 0.0078 | |
| SRP159526 | MBNL3 | 55796 | RNAseq | -1.6142 | 0.0000 | |
| SRP193095 | MBNL3 | 55796 | RNAseq | -1.4299 | 0.0000 | |
| SRP219564 | MBNL3 | 55796 | RNAseq | -0.9061 | 0.1205 | |
| TCGA | MBNL3 | 55796 | RNAseq | -0.2425 | 0.0611 |
Upregulated datasets: 0; Downregulated datasets: 6.
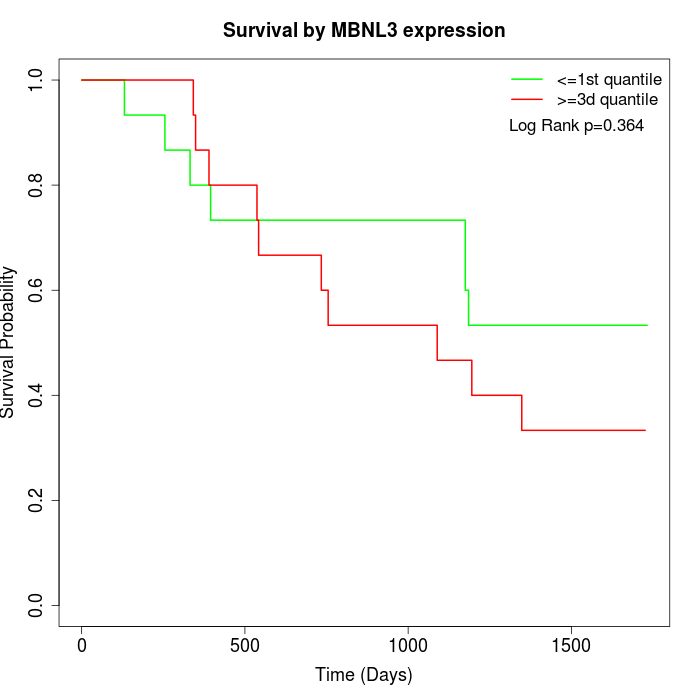
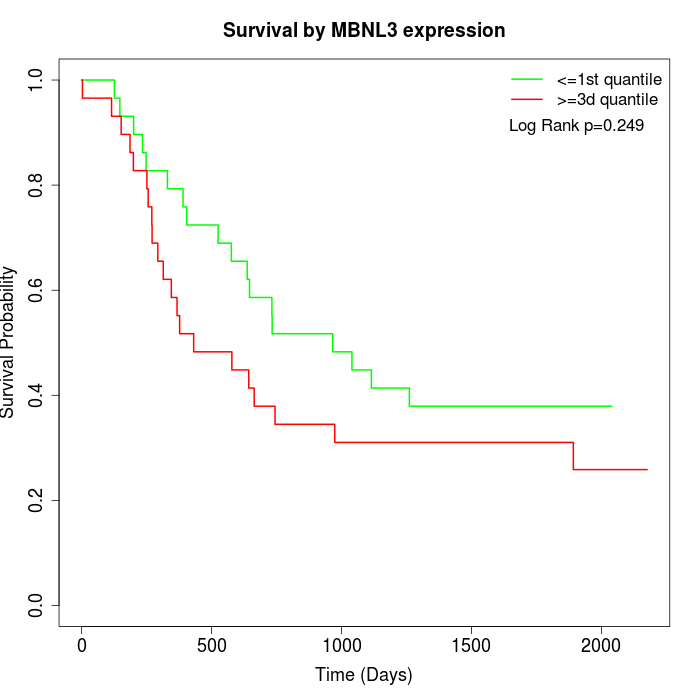
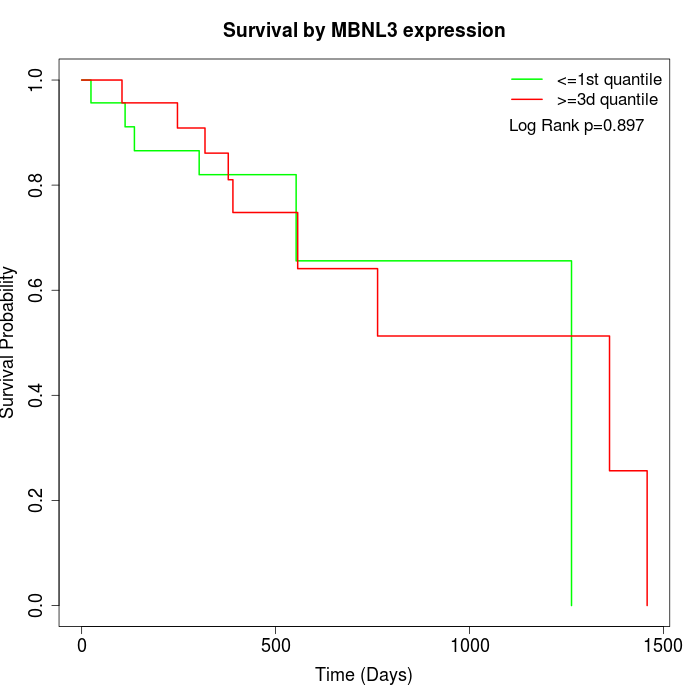
Survival by MBNL3 expression:
Note: Click image to view full size file.
Copy number change of MBNL3:
No record found for this gene.
Somatic mutations of MBNL3:
Generating mutation plots.
Highly correlated genes for MBNL3:
Showing top 20/1394 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MBNL3 | TSTD1 | 0.794989 | 3 | 0 | 3 |
| MBNL3 | OPHN1 | 0.780886 | 5 | 0 | 5 |
| MBNL3 | THAP2 | 0.78021 | 3 | 0 | 3 |
| MBNL3 | ZNF83 | 0.779976 | 3 | 0 | 3 |
| MBNL3 | C15orf48 | 0.775155 | 3 | 0 | 3 |
| MBNL3 | EPS15 | 0.774883 | 5 | 0 | 4 |
| MBNL3 | WDR77 | 0.772688 | 3 | 0 | 3 |
| MBNL3 | PINK1 | 0.762538 | 4 | 0 | 4 |
| MBNL3 | NDFIP2 | 0.759562 | 3 | 0 | 3 |
| MBNL3 | YAP1 | 0.756826 | 3 | 0 | 3 |
| MBNL3 | HMGN1 | 0.752717 | 4 | 0 | 4 |
| MBNL3 | ZNF300 | 0.75182 | 3 | 0 | 3 |
| MBNL3 | UBE2B | 0.751463 | 3 | 0 | 3 |
| MBNL3 | REPS1 | 0.750124 | 4 | 0 | 4 |
| MBNL3 | DNAJC1 | 0.748441 | 3 | 0 | 3 |
| MBNL3 | NDUFS4 | 0.747498 | 4 | 0 | 4 |
| MBNL3 | ABHD16A | 0.747462 | 3 | 0 | 3 |
| MBNL3 | MAL2 | 0.74504 | 4 | 0 | 4 |
| MBNL3 | STXBP5 | 0.744637 | 6 | 0 | 5 |
| MBNL3 | SGMS2 | 0.744006 | 3 | 0 | 3 |
For details and further investigation, click here