| Full name: mediator complex subunit 12 | Alias Symbol: CAGH45|HOPA|OPA1|TRAP230|KIAA0192|OKS | ||
| Type: protein-coding gene | Cytoband: Xq13.1 | ||
| Entrez ID: 9968 | HGNC ID: HGNC:11957 | Ensembl Gene: ENSG00000184634 | OMIM ID: 300188 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MED12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MED12 | 9968 | 216071_x_at | 0.3196 | 0.4600 | |
| GSE20347 | MED12 | 9968 | 216071_x_at | 0.0041 | 0.9669 | |
| GSE23400 | MED12 | 9968 | 211342_x_at | -0.0506 | 0.3521 | |
| GSE26886 | MED12 | 9968 | 211342_x_at | 0.2555 | 0.1084 | |
| GSE29001 | MED12 | 9968 | 211342_x_at | -0.0367 | 0.9090 | |
| GSE38129 | MED12 | 9968 | 211342_x_at | -0.0616 | 0.5018 | |
| GSE45670 | MED12 | 9968 | 216071_x_at | 0.0548 | 0.7084 | |
| GSE53622 | MED12 | 9968 | 33587 | 0.1409 | 0.0315 | |
| GSE53624 | MED12 | 9968 | 6252 | 0.0674 | 0.2052 | |
| GSE63941 | MED12 | 9968 | 216071_x_at | 0.3260 | 0.3796 | |
| GSE77861 | MED12 | 9968 | 211342_x_at | -0.0975 | 0.5776 | |
| GSE97050 | MED12 | 9968 | A_23_P73702 | 0.1194 | 0.5726 | |
| SRP007169 | MED12 | 9968 | RNAseq | 0.9527 | 0.0466 | |
| SRP008496 | MED12 | 9968 | RNAseq | 0.6360 | 0.0722 | |
| SRP064894 | MED12 | 9968 | RNAseq | 0.1331 | 0.5380 | |
| SRP133303 | MED12 | 9968 | RNAseq | -0.1075 | 0.4973 | |
| SRP159526 | MED12 | 9968 | RNAseq | 0.0735 | 0.8008 | |
| SRP193095 | MED12 | 9968 | RNAseq | 0.1557 | 0.1322 | |
| SRP219564 | MED12 | 9968 | RNAseq | 0.0474 | 0.8943 | |
| TCGA | MED12 | 9968 | RNAseq | -0.0290 | 0.5156 |
Upregulated datasets: 0; Downregulated datasets: 0.
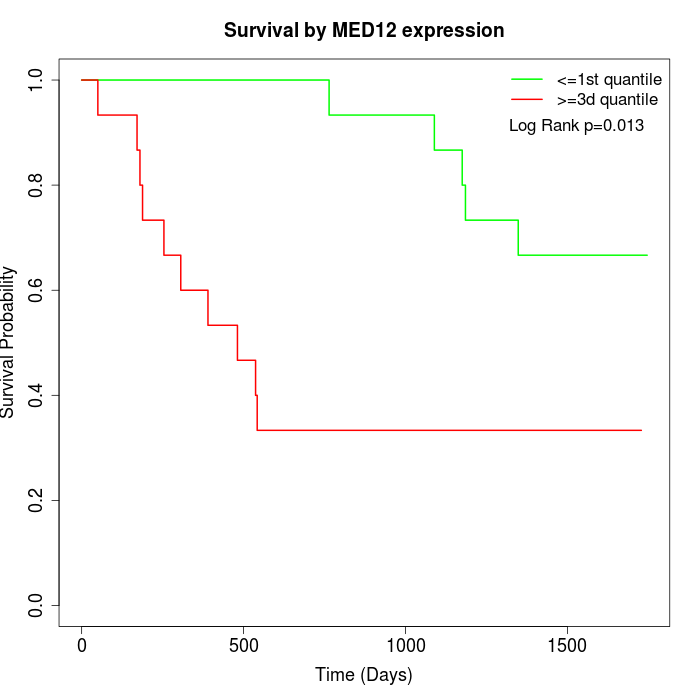
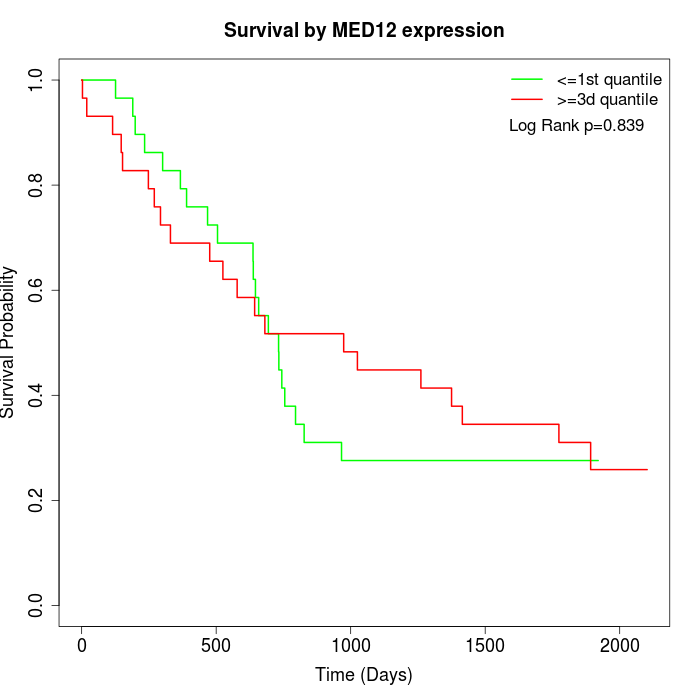
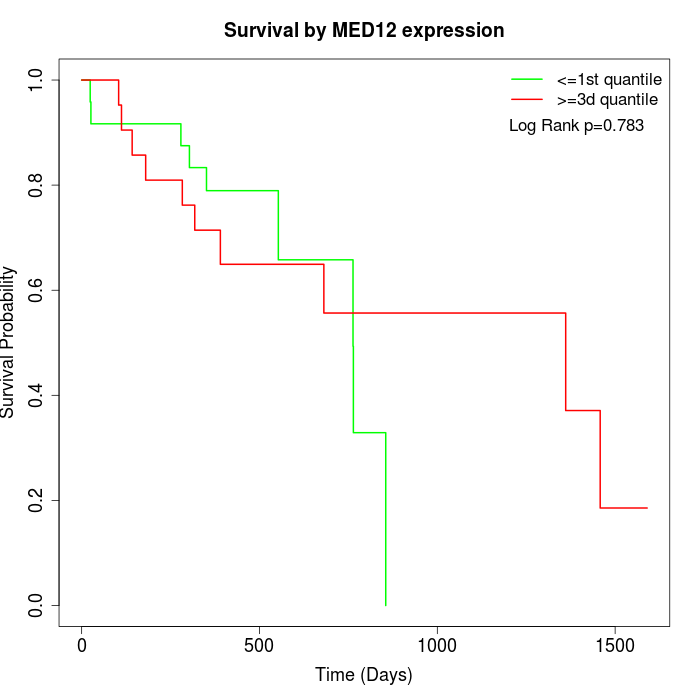
Survival by MED12 expression:
Note: Click image to view full size file.
Copy number change of MED12:
No record found for this gene.
Somatic mutations of MED12:
Generating mutation plots.
Highly correlated genes for MED12:
Showing top 20/279 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MED12 | PLD3 | 0.802682 | 3 | 0 | 3 |
| MED12 | ZNF830 | 0.794514 | 3 | 0 | 3 |
| MED12 | LY6E | 0.778452 | 3 | 0 | 3 |
| MED12 | TG | 0.765935 | 3 | 0 | 3 |
| MED12 | PIAS3 | 0.758555 | 4 | 0 | 4 |
| MED12 | SLFNL1 | 0.754027 | 3 | 0 | 3 |
| MED12 | PLEKHA4 | 0.743182 | 4 | 0 | 4 |
| MED12 | SPI1 | 0.73915 | 3 | 0 | 3 |
| MED12 | RNFT2 | 0.738774 | 3 | 0 | 3 |
| MED12 | ZDHHC14 | 0.732761 | 4 | 0 | 4 |
| MED12 | GDI1 | 0.727829 | 4 | 0 | 4 |
| MED12 | SLC25A29 | 0.727569 | 3 | 0 | 3 |
| MED12 | NUCB1 | 0.721383 | 4 | 0 | 4 |
| MED12 | REC8 | 0.717641 | 3 | 0 | 3 |
| MED12 | MAN2B1 | 0.712352 | 3 | 0 | 3 |
| MED12 | TNFRSF9 | 0.711249 | 4 | 0 | 4 |
| MED12 | COMT | 0.708735 | 3 | 0 | 3 |
| MED12 | SUGP1 | 0.707329 | 4 | 0 | 4 |
| MED12 | THBS4 | 0.706707 | 3 | 0 | 3 |
| MED12 | POLR1C | 0.705689 | 3 | 0 | 3 |
For details and further investigation, click here