| Full name: schlafen like 1 | Alias Symbol: FLJ23878 | ||
| Type: protein-coding gene | Cytoband: 1p34.2 | ||
| Entrez ID: 200172 | HGNC ID: HGNC:26313 | Ensembl Gene: ENSG00000171790 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SLFNL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SLFNL1 | 200172 | 1554074_s_at | 0.2148 | 0.3743 | |
| GSE26886 | SLFNL1 | 200172 | 1554074_s_at | -0.1895 | 0.0602 | |
| GSE45670 | SLFNL1 | 200172 | 1554074_s_at | 0.0125 | 0.8891 | |
| GSE53622 | SLFNL1 | 200172 | 6555 | 0.0185 | 0.8161 | |
| GSE53624 | SLFNL1 | 200172 | 6555 | 0.1553 | 0.0174 | |
| GSE63941 | SLFNL1 | 200172 | 1554074_s_at | 0.1476 | 0.3349 | |
| GSE77861 | SLFNL1 | 200172 | 1554074_s_at | -0.0312 | 0.8473 | |
| GSE97050 | SLFNL1 | 200172 | A_23_P389141 | -0.0433 | 0.8728 | |
| SRP133303 | SLFNL1 | 200172 | RNAseq | 0.5629 | 0.1189 | |
| TCGA | SLFNL1 | 200172 | RNAseq | 0.1037 | 0.6501 |
Upregulated datasets: 0; Downregulated datasets: 0.
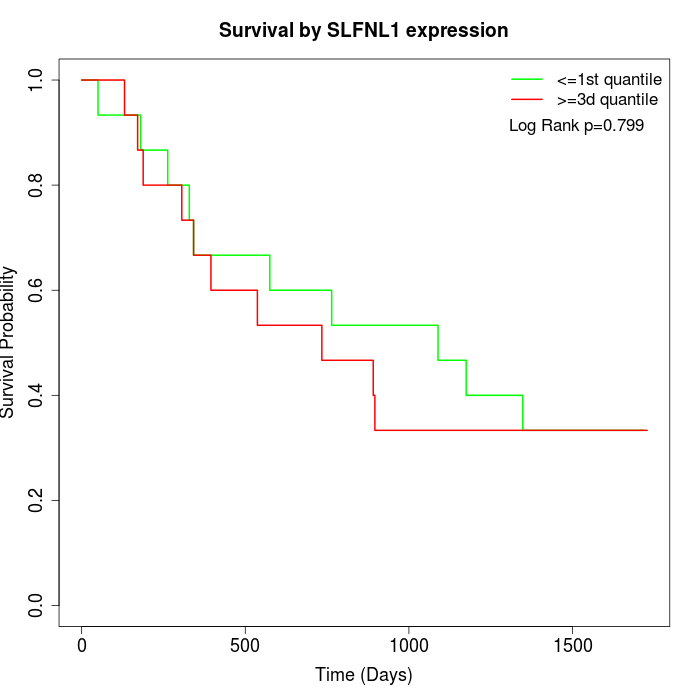
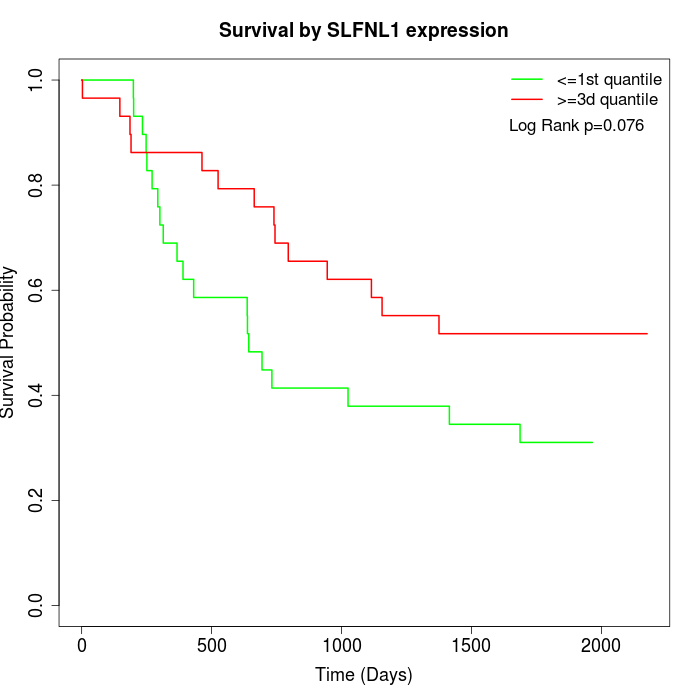
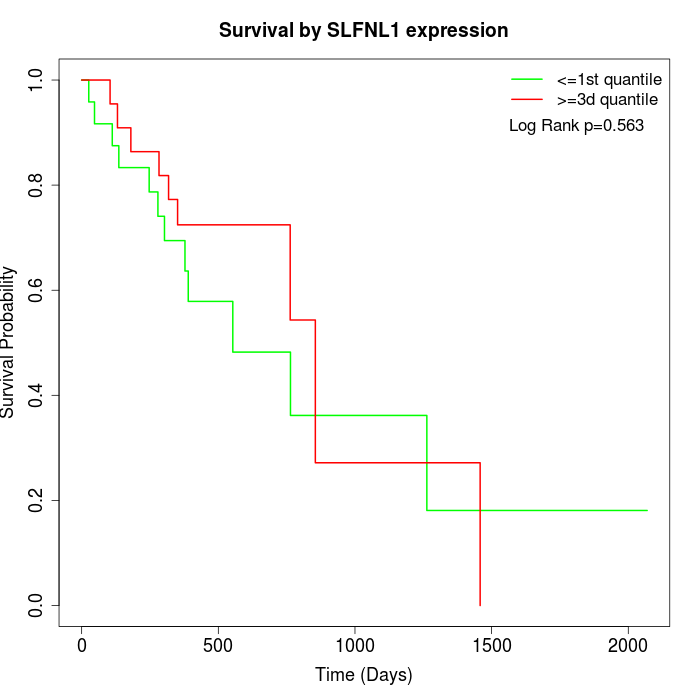
Survival by SLFNL1 expression:
Note: Click image to view full size file.
Copy number change of SLFNL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SLFNL1 | 200172 | 3 | 4 | 23 | |
| GSE20123 | SLFNL1 | 200172 | 2 | 3 | 25 | |
| GSE43470 | SLFNL1 | 200172 | 7 | 2 | 34 | |
| GSE46452 | SLFNL1 | 200172 | 3 | 1 | 55 | |
| GSE47630 | SLFNL1 | 200172 | 9 | 2 | 29 | |
| GSE54993 | SLFNL1 | 200172 | 0 | 1 | 69 | |
| GSE54994 | SLFNL1 | 200172 | 13 | 2 | 38 | |
| GSE60625 | SLFNL1 | 200172 | 0 | 0 | 11 | |
| GSE74703 | SLFNL1 | 200172 | 6 | 1 | 29 | |
| GSE74704 | SLFNL1 | 200172 | 1 | 0 | 19 | |
| TCGA | SLFNL1 | 200172 | 16 | 16 | 64 |
Total number of gains: 60; Total number of losses: 32; Total Number of normals: 396.
Somatic mutations of SLFNL1:
Generating mutation plots.
Highly correlated genes for SLFNL1:
Showing top 20/61 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SLFNL1 | SLC39A3 | 0.820125 | 3 | 0 | 3 |
| SLFNL1 | MED12 | 0.754027 | 3 | 0 | 3 |
| SLFNL1 | XYLB | 0.748425 | 3 | 0 | 3 |
| SLFNL1 | PTPN7 | 0.74663 | 3 | 0 | 3 |
| SLFNL1 | ZNF419 | 0.732389 | 3 | 0 | 3 |
| SLFNL1 | CYB561 | 0.732035 | 3 | 0 | 3 |
| SLFNL1 | CUL7 | 0.723146 | 3 | 0 | 3 |
| SLFNL1 | PIK3CB | 0.681606 | 3 | 0 | 3 |
| SLFNL1 | MLLT6 | 0.680974 | 3 | 0 | 3 |
| SLFNL1 | SIDT1 | 0.670469 | 3 | 0 | 3 |
| SLFNL1 | CERS6 | 0.667337 | 3 | 0 | 3 |
| SLFNL1 | ACSL1 | 0.653361 | 3 | 0 | 3 |
| SLFNL1 | DBP | 0.6501 | 3 | 0 | 3 |
| SLFNL1 | SCML4 | 0.648754 | 4 | 0 | 3 |
| SLFNL1 | SETD5 | 0.64455 | 4 | 0 | 3 |
| SLFNL1 | MX1 | 0.644542 | 3 | 0 | 3 |
| SLFNL1 | CD3E | 0.641946 | 3 | 0 | 3 |
| SLFNL1 | MPV17L | 0.641665 | 4 | 0 | 3 |
| SLFNL1 | XRN1 | 0.639146 | 4 | 0 | 3 |
| SLFNL1 | GATA1 | 0.629211 | 4 | 0 | 3 |
For details and further investigation, click here